New structures show how HPF1 and PARP form a ‘composite’ enzyme to initiate the response to DNA damage
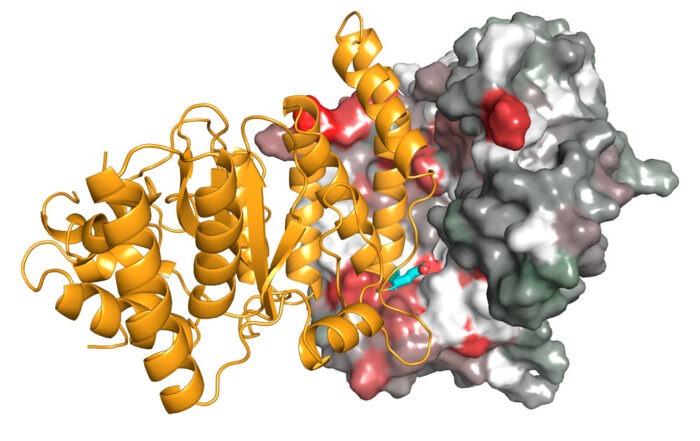
Detection of DNA damage requires a quick response and dynamic regulation of proteins. Better understanding how DNA repair pathways are initiated could have great clinical implications, particularly for cancer therapy. A team of scientists, including members of David Neuhaus’ group at the LMB, led by Ivan Ahel at the University of Oxford, has now visualised the interaction between two proteins, PARP and HPF1, to show how they work together to initiate DNA repair.
PARPs (poly (ADP-ribose) polymerases) drive a kind of protein modification reaction called ADP-ribosylation in response to detection of DNA damage. This modification is used to regulate a plethora of proteins, ultimately leading to efficient repair of the damage. Although PARPs have been well-known and widely studied for decades, and are even targets for approved cancer drugs, many aspects of their mechanism of action had eluded scientists and even the nature of sites of ADP-ribosylation in response to DNA damage detection was unclear.
In a series of studies over the last 4 years, Ivan Ahel’s group at the Sir William Dunn School of Pathology at the University of Oxford demonstrated for the first time that the main target of ADP-ribosylation, in living cells in the context of genotoxic stress, were protein serine residues. In addition, they found that an accessory factor called HPF1 was needed alongside a PARP enzyme (PARP1 or PARP2) for this to happen. In isolation, PARP1 and PARP2 modify glutamate or aspartate amino acid side chains, implying that HPF1 causes a specificity switch, redirecting these enzymes towards the serine sites that orchestrate DNA repair.
Using X-ray crystallography, Ivan’s group has now determined structures of HPF1 and its complex with PARP2. To crystallise the complex, one part of the PARP catalytic domain had to be deleted in order to stabilise the interaction with HPF1. David’s group, in the LMB’s Structural Studies Division, was then able to use NMR spectroscopy, a technique that works well for characterising weak interactions in solution, to study the HPF1-PARP1 complex with this part of the PARP catalytic domain included and visualise a similar interaction as seen in the crystal.
© Marcin Suskiewicz, Oxford University
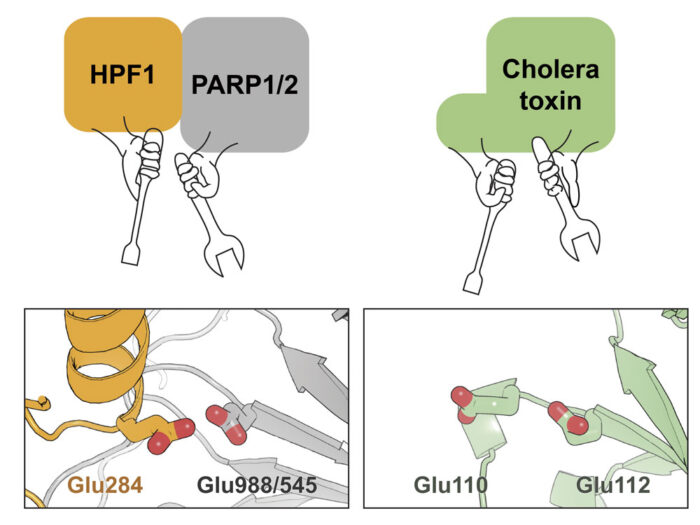
By analysing these structures, as well as other data relating to the function of this complex, the team demonstrated that both PARP and HPF1 contribute to the chemical “tools” in the active site that catalyses the ADP-ribosylation on serine side chains. Interestingly, although this active site is made by combining two proteins, it resembles the arginine ADP-ribosylation active site in cholera toxin, which is just a single molecule. This is presumably an example of convergent evolution with a similar functional outcome being achieved through two different routes.
PARP inhibitors are of increasing importance in cancer treatment and have been approved for treatment of certain types of advanced ovarian and breast cancers. They are also being trialled for use in prostate and colon cancer, and even in stroke, heart disease, and neurodegenerative diseases. These new structures, and the concomitant increased understanding of how PARPs create the ADP-ribosylation that initiates DNA repair processes, could potentially lead to improvements in PARP inhibitor efficacy and novel types of inhibitors for treatment of this wide range of diseases.
The work was funded by the MRC, Wellcome Trust, BBSRC, Cancer Research UK, EMBO, and an LMB/AstraZeneca BlueSkies postdoctoral fellowship.
Further references:
HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation. Suskiewicz, MJ., Zobel, F., Ogden, TEH., Fontana, P., Ariza, A., Yang, JC., Zhu, K., Bracken, L., Hawthorne, WJ., Ahel, D., Neuhaus, D., Ahel, I. Nature (Epub ahead of print)
David’s group page
Ivan Ahel’s page