 Mitochondria are organelles within eukaryotic cells that likely evolved from an ancient bacterium that was engulfed by a primordial eukaryote. Within mitochondria, mitochondrial ribosomes (mitoribosomes) synthesise a subset of essential proteins encoded by the mitochondrial genome. Although mitoribosomes share a common ancestor with bacterial ribosomes, they have undergone substantial changes during evolution that have resulted in morphologically diverse mitoribosomes across the eukaryotic kingdom.
Mitochondria are organelles within eukaryotic cells that likely evolved from an ancient bacterium that was engulfed by a primordial eukaryote. Within mitochondria, mitochondrial ribosomes (mitoribosomes) synthesise a subset of essential proteins encoded by the mitochondrial genome. Although mitoribosomes share a common ancestor with bacterial ribosomes, they have undergone substantial changes during evolution that have resulted in morphologically diverse mitoribosomes across the eukaryotic kingdom.
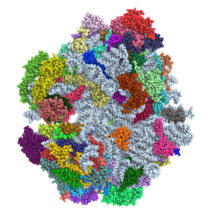
New research from Venki Ramakrishnan’s group in the LMB’s Structural Studies Division has revealed the complete structure of the mitoribosome from baker’s yeast (Saccharomyces cerevisiae). This is the first near atomic-resolution structure of a non-mammalian mitoribosome and expands upon an earlier study by the group into the large subunit of the yeast mitoribosome. Lead-author Nirupa Desai together with Alexey Amunts purified the mitoribosomes directly from yeast cells grown in the lab and visualised them using cryo-electron microscopy. Nirupa and Alan Brown then interpreted these images to build a model that revealed the atomic details of the 75 different RNA and protein components within the yeast mitoribosome.
Amazingly, despite having a shared ancestor, the yeast mitoribosome has 27 additional proteins compared to the bacterial ribosome (14 of which are in the newly resolved small subunit). The structure of the yeast mitoribosome is also very different from the human mitoribosome (solved by Venki’s group in 2015) and the porcine mitoribosome (solved by Nenad Ban’s group in 2015). Unlike these mammalian mitoribosomes the yeast mitoribosome has many new sections of rRNA and many of the proteins differ too. The task is now to identify why these differences arose and what role the new proteins are playing in the yeast mitoribosome. The structure already provides clues. For example, some of the proteins form a unique V-shaped canyon at the exit to the mRNA channel that may accommodate mRNAs and bind protein translational activators that control translation in yeast but not in mammalian mitochondria.
The structure of the yeast mitoribosome is a particularly valuable resource, as S. cerevisiae remains the model organism to study mitochondrial translation due to tools to modify its mitochondrial genome. The structure also reveals new insights into mitoribosomal evolution, the species-specific mechanisms that control mitochondrial translation, and novel adaptations that facilitate the optimal translation of the few genes encoded by the mitochondrial genome. The structure should act as reference to explore further the intimate relationship between ribosome structure and protein synthesis and how this relationship changes under different evolutionary pressures.
This work was funded by the MRC, the Wellcome Trust, the Agouron Institute and the Louis-Jeantet Institute.
Further references:
Paper in ScienceVenki’s group page
Previous Insight on Research article: Breakthrough in structural biology reveals mitochondrial ribosome structure
The structure of the human mitochondrial ribosome
The complete structure of the 55S mammalian mitochondrial ribosome