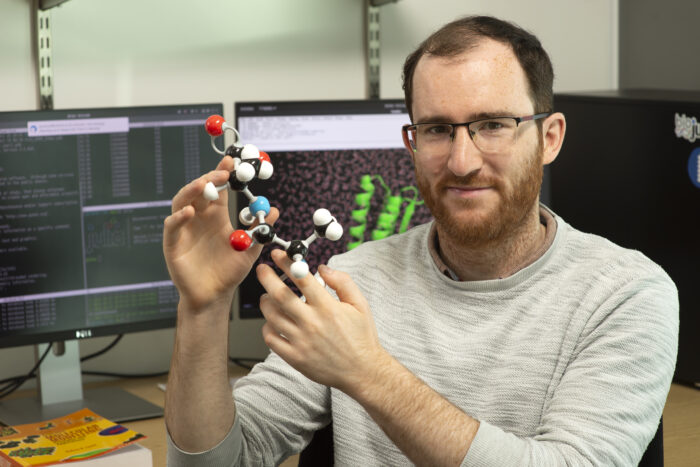
The LMB is delighted to announce Joe Greener as the newest Group Leader in our Structural Studies Division, where he will develop differentiable molecular simulation methods to improve biomolecular force fields.
Joe commented, “The explosion of biological data, improvements in computing and exciting machine learning advances mean that now is an exciting time to be developing methods for molecular simulation. I am looking forward to starting a research group at the LMB among such talented scientists.”
Force fields are a computational method used to aid molecular modelling by estimating the forces between atoms both within and between molecules. Using force fields, molecular dynamics has proven successful in providing biological insights through mechanistic interpretations of experimental data and suggestions for future studies. However, the force fields used to interpret how atoms interact rely on parameters skewed towards the notion that proteins are folded and, as such, fail when applied to disordered or aggregated proteins commonly found in disease. Development of new force fields with optimised parameters to study disordered proteins is a challenging and time-consuming process, particularly when trying to adapt multiple parameters at once.
Differentiable molecular simulation (DMS) tackles this by using a simulation to obtain gradients that indicate how each parameter affects the accuracy, the data from which can then be harnessed to improve the force field. Joe’s research has already illustrated the potential for DMS by developing a force field that can predict the folding of small proteins. With further research, he has shown the scope for an all-atom force field that works across a range of biomolecular systems. This approach holds significant appeal as a large number of parameters are able to be improved simultaneously. Joe plans to further apply his improved DMS approach to better understand aggregated proteins found within many neurodegenerative diseases.
After receiving an MSci and BA Hons in Natural Sciences (Chemistry) from Emmanuel College, University of Cambridge, Joe went on to an MSc in Bioinformatics and Theoretical Systems Biology and then a PhD in Structural Bioinformatics, both at Imperial College London. Following this, Joe undertook postdoctoral studies in the lab of David Jones at University College London and the Francis Crick Institute, before joining the LMB as a Senior Investigator Scientist in September 2021. For the past two years he has been developing methods to improve implicit solvent force fields.