Scientists use single cell RNA sequencing data to generate the world’s first healthy human kidney cell atlas, providing a critical stepping-stone to understanding the pathways that lead to kidney disease and help identify future therapies
Chronic kidney disease affects more than 500 million patients worldwide, but there are no specific treatments that prevent disease progression. A lack of tissue availability has limited research into human kidney function and animal research hasn’t translated into treatments for human patients. Menna Clatworthy’s group, in the University of Cambridge Molecular Immunity Unit, which is housed within the LMB, has now generated the first atlas-scale dataset map of human kidney cells, which will begin to address this knowledge gap.
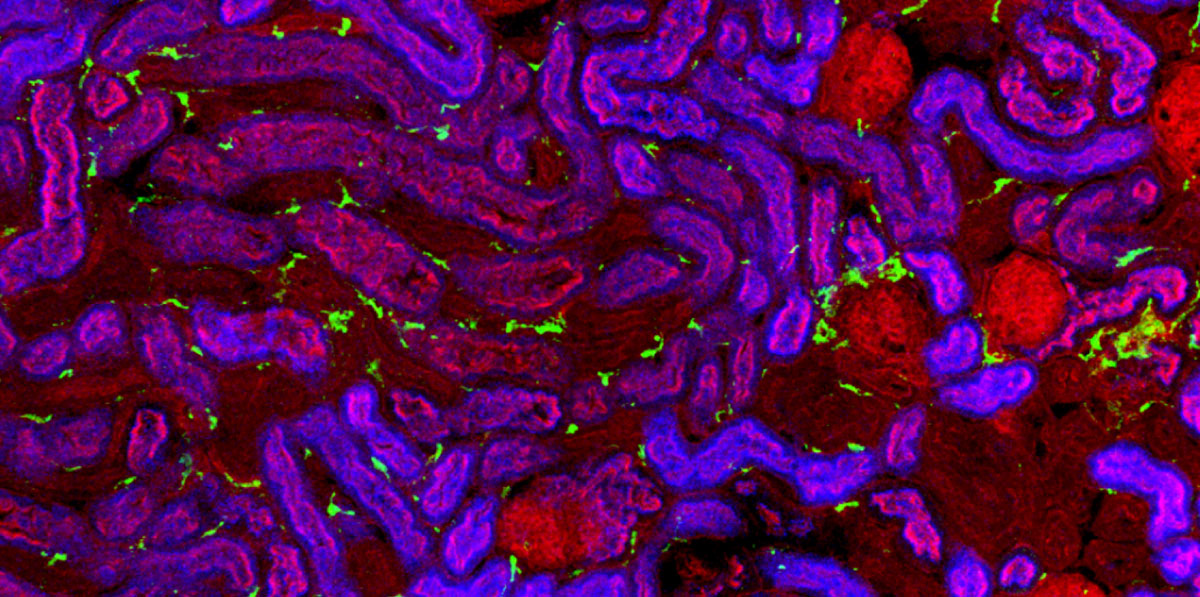
Kidneys are made up of a variety of different cells that interact to deliver normal function, but the exact composition and organisation of these cells is not known. Using single cell RNA sequencing, Benjamin Stewart and John Ferdinand from Menna’s group, in collaboration with Sam Behjati’s and Sarah Teichmann’s groups at the Wellcome Sanger Institute and Muzlifah Haniffa’s group at Newcastle University, have investigated human kidney cells across the human lifespan.
What is single cell RNA sequencing?
By sequencing RNA molecules in cells, scientists can observe which genes are being expressed in those cells. Standard methods of doing this have involved analysis of large populations of cells. However, this may hide rare cell-types and mask small differences between different individual cells.
Single cell RNA sequencing provides gene expression data for individual cells, arming researchers with a deeper understanding of the diversity of cells present in a tissue or organ.
By performing single cell RNA sequencing on over 65,000 cells from fetal and adult kidneys, the team was able to investigate the spatial and temporal organisation of different cell-types across the kidney and produce a map that will form the backbone of the first version of the human kidney cell atlas in the Human Cell Atlas project.
One surprising observation was that there were many immune cells in the healthy kidney and even in the developing kidney long before birth. Comparing fetal and adult kidneys, Benjamin and John were also able to show how the epithelial cells that line kidney tubules change and are able to contribute to immune function postnatally. On top of this, the team characterised the molecular signals used by the epithelial cells to direct immune cells to the specific regions where they are most needed to defend the kidney from infection.
This study will transform our understanding of the cellular communication networks in the human kidney and, importantly, can pinpoint which cells express kidney disease-associated genes. Knowledge of which immune cells are present in different parts of the kidney will also impact our understanding of how to treat infectious diseases and autoimmune kidney diseases, both of which are important causes of kidney failure.
A full map of the cells that make up the kidney will also be useful for researchers who want to grow kidney organoids in order to test drugs or study kidney development, and might even be useful for scientists growing kidneys for transplant. The development of future therapies for kidney disease could have huge implications for cost saving in comparison with dialysis and improvement in the quality of life of patients.
The work was funded by the MRC, Wellcome Trust, CRUK, NIHR, Arthritis Research UK, St Baldrick’s Foundation, Lister Institute for Preventative Medicine, International Society for Advancement of Cytometry, Kidney Research UK, EMBO, Human Frontier Science Program, Children with Cancer UK, and Cambridge Biomedical Campus.
Further references:
Spatio-temporal immune zonation of the human kidney. Stewart, BJ., Ferdinand, JR., Young, MD., Mitchell, TJ., Loudon, KW., Riding, AM., Richoz, N., Frazer, GL., Staniforth, JUL., Vieira Braga, FA., Botting, RA., Popescu, D-M., Vento-Tormo, R., Stephenson, E., Cagan, A., Farndon, SJ., Polanski, K., Efremova, M., Green, K., Del Castillo Velasco-Herrera, M., Guzzo, C., Collord, G., Mamanova, L., Aho, T., Armitage, JN., Riddick, ACP., Mushtaq, I., Farrell, S., Rampling, D., Nicholson, J., Filby, A., Burge, J., Lisgo, S., Lindsay, S., Bajenoff, M., Warren, AY., Stewart, GD., Sebire, N., Coleman, N., Haniffa, M., Teichmann, SA., Behjati, S., Clatworthy, MR. Science 365(6460): 1461-1466
Menna’s group page
Sam Behjati’s group page
Muzlifah Haniffa’s group page
Sarah Teichmann’s group page
Human Kidney Cell Atlas