During eukaryotic cell division (mitosis) the cell’s chromosomes are duplicated and then equally separated into two new daughter cells. To prevent errors in mitosis cells employ checkpoints that monitor and coordinate the correct order of events. Checkpoints either delay cell division, or if unrecoverable, cause cell death. For the first time, David Barford’s group in the LMB’s Structural Studies Division, have determined the molecular mechanism by which the checkpoint signal in mitosis inhibits a large protein complex (the APC/C) that regulates chromosome separation.
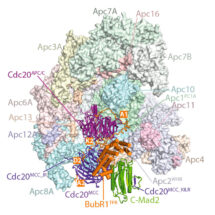
The separation of sister chromosomes in mitosis is driven by the mitotic spindle. Chromosomes attach to the spindle through their kinetochores. In the dividing eukaryotic cell the spindle assembly checkpoint (SAC) ensures each daughter cell inherits an identical set of chromosomes. The SAC coordinates the correct attachment of kinetochores to the mitotic spindle with activation of the anaphase-promoting complex/cyclosome (APC/C), a multi-protein complex responsible for initiating chromosome separation. In response to unattached kinetochores, the SAC generates the mitotic checkpoint complex (MCC), a multimeric assembly that inhibits the APC/C, delaying chromosome segregation. Using cryo-electron microscopy, Claudio Alfieri and Leifu Chang, in David’s group, have determined the near-atomic resolution structure of an APC/CMCC complex. They also revealed how degron-like sequences of a MCC subunit (BubR1) block degron recognition sites on Cdc20, the APC/C coactivator subunit (Cdc20APC/C) responsible for substrate interactions. BubR1 also obstructs binding of UbcH10 (APC/C’s initiating E2) to repress APC/C ubiquitination activity. Conformational variability of the complex allows for UbcH10 association, and they show from a structure of APC/CMCC in complex with UbcH10 how the Cdc20 subunit intrinsic to the MCC (Cdc20MCC) is ubiquitinated, a process that results in spontaneous reactivation of APC/C when the checkpoint signal is terminated.
Errors in this process during mitosis and meiosis cause an unequal distribution of chromosomes (aneuploidy) that underlies genetic diseases such as cancer and developmental and birth defects. This study has shown the basis for inhibiting the APC/CMCC complex. This may have future use in development of treating tumours, where blocking mitosis could be used to block cell proliferation and promote programmed cell death
The work was funded by the MRC and Cancer Research UK.
Further references:
David’s group page
Paper in Nature
Nature News and Views
Previous related paper: Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature 533: 260-264