 Researchers in Greg Jefferis’s group in the LMB’s Neurobiology Division have developed a new online tool to analyse images of neurons. This tool, known as NBLAST, is free and available to all. NBLAST enables researchers to measure the similarity between neurons and organise them into neuron families, akin to tools such as BLAST that allow protein sequences to be compared.
Researchers in Greg Jefferis’s group in the LMB’s Neurobiology Division have developed a new online tool to analyse images of neurons. This tool, known as NBLAST, is free and available to all. NBLAST enables researchers to measure the similarity between neurons and organise them into neuron families, akin to tools such as BLAST that allow protein sequences to be compared.
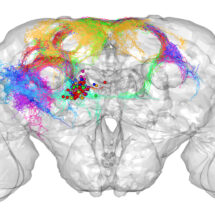
Neuroscience is seeing a period of major growth in the structural characterisation of neurons. Every neuron type has its own unique shape and location within the brain, and its own set of unique connections to other neurons, sensory or effector cells. Knowing the circuit organisation and connectivity of neurons enables researchers to understand how they generate and control different behaviours.
The pattern of neuronal connections is the same within all members of the same species and is known as a connectome. The first connectome, the set of all the connections between 302 neurons of the nematode C. elegans, was described by the LMB in the 1970s and early ‘80s. Now there are well-advanced efforts to obtain connectomes of the fruit fly and one day, the mouse.
Using a data set of 16,000 fruit fly neurons generously shared by the group of Professor Ann-Shyn Chiang in Taiwan, Greg and Marta Costa designed and validated the algorithm used in NBLAST. The software and search tools were further refined by Greg and James Manton. NBLAST works by comparing the shape and position of neurons to produce a similarity score, which can then used to identify similar neurons and organise neurons into families. Importantly, NBLAST is fast as well as sensitive, taking only 2 milliseconds to complete one comparison – a database of 16,000 neurons can be searched in 30 seconds.
NBLAST is already proving useful in fly brain research to identify neuron types, organize large datasets and to identify new subtypes of neurons that have not previously been described. NBLAST is also known to work for other model species such as zebrafish. Finally, NBLAST can analyse neurons traced from electron microscope data; early results from Greg’s group and collaborators at HHMI Janelia Research Campus suggest that it will be an important tool in ongoing work to determine a complete fruit fly connectome.
The work was funded by the MRC, ERC and EMBO Young Investigator Programme.