Phenogenomic analysis – a combination of phenotyping and whole genome sequencing – reveals insights into bacterial pathobiology
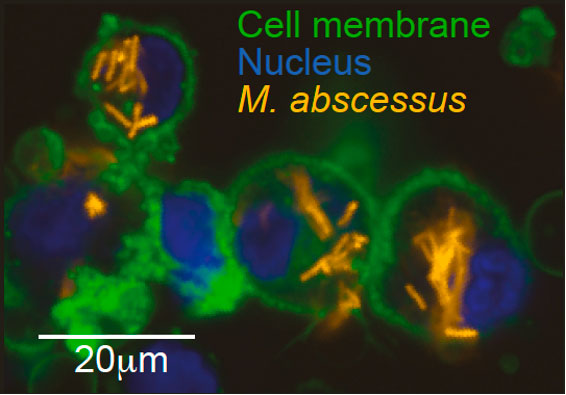
Mycobacterium abscessus (M. abscessus) is a multidrug-resistant species of nontuberculous mycobacteria (NTM) which has rapidly grown over the last two decades and poses a major threat to individuals with Cystic Fibrosis and other chronic lung diseases. Medical and scientific responses to emerging and established pathogens is often hindered by a lack of knowledge of the genetic determinants of factors such as virulence, drug resistance and clinical outcomes, which could be used to forecast patient trajectories and identify possible treatments. Andres Floto’s group from the University of Cambridge’s Molecular Immunity Unit, which is housed at the LMB, have now illustrated how combining experimental phenotyping with whole genome sequencing can provide a phenogenomic map of M. abscessus which reveals actionable, systems-level insights into bacterial pathobiology.
Lead author Lucas Boeck (formerly in Andres’ group, now Group Leader at the University of Basel) analysed the behaviour of hundreds of clinical isolates of M. abscessus. He discovered three distinct clusters of isolates, each with different virulence traits and associated with a different clinical outcome. It is likely each of these isolate clusters represents a different evolutionary trajectory taken by M. abscessus during host adaption.
In undertaking this study, the group combined genome-wide association studies (GWAS) with proteome-wide computational structural modelling to identify likely causal genetic variants. Direct coupling analysis (DCA) was then used to determine co-evolving, and therefore potentially interacting networks between genes. These approaches allowed the group to build a network of genes involved in clinically significant changes in virulence. To validate these findings of M. abscessus virulence factors, they used in vivo CRISPR-based gene silencing methods developed by group member Sophie Burbaud. Ultimately this showed that the clinical outcome of patients infected with M. abscessus was related to the presence of particular mutations identified by this new ‘phenogenomic’ approach.
These findings provide proof of concept for the group’s phenogenomic approach to understanding new pathogens. Their phenogemoic map of M. abscessus could help to define critical pathways involved in virulence and drug resistance. Moreover, these results demonstrate the power of combining genetic analysis with proteome-wide structural modelling as a method for understanding how gene mutations cause changes in cell outcomes which, although the group focused on bacteria in this instance, could be applied further to wider genomic datasets.
This work was funded by The Wellcome Trust, The UK Cystic Fibrosis Trust, Vertex, NIHR, The Botnar Foundation, The Swiss National Science Foundation, The Bangerter-Rhyner and Helmut Horten Foundation, European Respiratory Society, EMBO and The German Research Foundation (DFG).
Further references
Mycobacterium abscessus pathogenesis identified by phenogenomic analyses. Boeck, L., Burbaud, S., Skwark, M., Pearson, WH., Sangen, J., Wuest, AW., Marshall, EKP., Weimann, A., Everall, I., Bryant, JM., Malhotra, S., Bannerman, BP., Kierdorf, K., Blundell, TL., Dionne, MS., Parkhill, J., Floto, RA. Nature Microbiology
Andres’ group page
Lucas Boeck’s page