 One of the most commonly mutated genes in human cancers is the lipid kinase PIK3CA (phosphoinositide 3-kinase alpha). Mutations can activate this enzyme, resulting in proliferation of tumour cells and resistance to programmed cell death or apoptosis.
One of the most commonly mutated genes in human cancers is the lipid kinase PIK3CA (phosphoinositide 3-kinase alpha). Mutations can activate this enzyme, resulting in proliferation of tumour cells and resistance to programmed cell death or apoptosis.
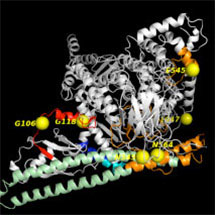
By applying the powerful technique of hydrogen/deuterium exchange mass spectrometry to map this complex system, a team of researchers in Roger Williams group in the LMB’s PNAC Division have succeeded in providing new insights into how mutations of the PIK3CA affect its function.
An article, published in PNAS, sets out the team’s findings. As lead researcher John Burke explains: “Using this technique we were able to show that the enzyme undergoes a complex change in its shape – from a closed, inhibited form free in the cell to an open, activated form on the membrane surrounding the cell. Mutations scattered throughout the structure cause changes in conformation that mimic dynamic events that happen in the natural process of activation of the enzyme.”
The conformational changes that are associated with activation of the enzyme on membranes include opening and closing of pockets far removed from the active site of the enzyme. By targeting these pockets, it may be possible to develop therapeutic inhibitors of the enzyme that would be specific for the oncogenic mutants.
The work was funded in part by the Medical Research Council, British Heart Foundation, EMBO and the Swiss National Science Foundation.