Correlative microscopy, in situ imaging of protein organisation and yeast genetics refine our understanding of ER-plasma membrane contact sites
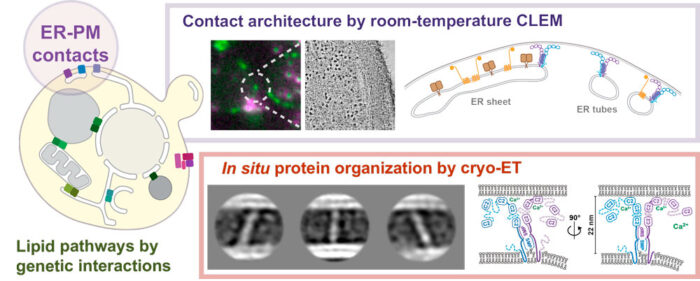
By combining fluorescence microscopy and electron tomography, Wanda Kukulski’s lab in Cell Biology Division has visualised protein structures that bridge contact sites between the endoplasmic reticulum and plasma membrane in yeast, in their native environment i.e. within the cell. Using this, the scientists have proposed a model on how tricalbins – proteins that facilitate lipid transport between organelles – are arranged because this organisation is critical to how cells maintain asymmetric distribution of different types of lipids. This uneven distribution assists in lipid flux.
They have also found that contact sites are more diverse in their composition than previously thought, and that membrane morphology correlates with composition. And finally, this study suggests that lipid distribution in cells is robust because the routes for lipid fluxes are highly redundant. “This makes sense given how fundamental lipids are for the identity and function of various cellular membranes,” says Wanda. Contact sites are important for lipid homeostasis and metabolism, which are key elements to maintain healthy organism physiology. Dysfunctions in lipid homeostasis can cause diseases, such as, diabetes, obesity and heart conditions.
Logistics of a cell
Though cells are the smallest unit of living things, they have many compartments within themselves, the organelles. The organelles need to communicate and transport material, such as lipids, to each other – therefore, there is a complex transport system. While some of these traffic routes are well understood, others, such as the communication at contact sites, have remained elusive. This is in part because there is little knowledge about the architecture of membrane contact sites, such as, between endoplasmic reticulum and plasma membranes (ER-PM).
Knowledge of the architecture of proteins at the ER-PM contact site (see graphic) is key to understanding how processes at contact sites occur, how important the precise organisation of the sites is, and how lipid fluxes within the network of contact sites are coordinated. These contact sites are key elements of how a cell is organised, and therefore, understanding how bridging proteins function is important.
Early steps in visualising proteins in situ
We have little information about this architecture in part because until recently, we did not have the imaging tools to identify and visualise cellular components at sufficiently high resolution. A second reason is that the network of lipid fluxes has many redundant routes, which makes it difficult to study them. But yeast genetics are a powerful way to unfold such redundancies.
“There is still a lot to discover on how the logistics in a cell function, and how the spatial architecture of cells contributes to it,” says Wanda Kukulski.
Patrick Hoffmann of Wanda’s Lab, supported by Tanmay Bharat, Group Leader at Oxford University, gained structural information by using averaging methods on protein particles found in electron cryo-tomograms of yeast cells. For this, the scientists used a focused ion beam to remove material from vitrified cells until the cells were thin enough for electron cryo-tomography (cryo-ET). Cryo-ET is an imaging technique used to produce high-resolution 3D views of samples, typically of cells.
Meanwhile, supported by Liz Miller, Group Leader in Cell Biology Division, Patrick used the power of yeast genetics in various ways, in particular by combining tricalbin gene deletion mutants with other gene deletions. Doing this systematically in what is known as high-content screens, allowed to identify genes that encode proteins, and sometimes components of complete pathways, which have overlapping functions with tricalbins. This approach revealed that tricalbin function is intricately embedded into cellular lipid fluxes.
This paper reflects the broad interest of cell and structural biologists to visualise protein structures in their native environment, that is, the cell. In the case of contact site bridging and lipid transfer proteins, this is key to understand how they work. “The very fact that we have the possibility to do in situ structural biology, even in its infancy, is absolutely thrilling,” adds Wanda. But there is still a long way to go since the current state of technology doesn’t provide the resolution required to understand the molecular mechanisms yet.
This work was supported by the MRC, the Wellcome Trust, the Royal Society and the Vallee Foundation.
Further references
Tricalbins contribute to cellular lipid flux and form curved ER-PM contacts that are bridged by rod-shaped structures. Hoffmann, PC., Bharat, TAM., Wozny, MR., Boulanger, J., Miller, EA., Kukulski, W. Developmental Cell 51(4)
Wanda Kukulski’s Group Page
Liz Miller’s Group Page
Tanmay Bharat’s page