 The spliceosome is a molecular machine, which together with RNA polymerases and ribosomes plays a critical role in basic gene expression. Research by Kiyoshi Nagai’s group in the LMB’s Structural Studies Division, has previously revealed the structure of the spliceosome in a fully active, substrate-bound state, immediately after its first catalytic reaction. The group has now expanded upon this work revealing the near-atomic level structure of the spliceosome just before mRNA formation.
The spliceosome is a molecular machine, which together with RNA polymerases and ribosomes plays a critical role in basic gene expression. Research by Kiyoshi Nagai’s group in the LMB’s Structural Studies Division, has previously revealed the structure of the spliceosome in a fully active, substrate-bound state, immediately after its first catalytic reaction. The group has now expanded upon this work revealing the near-atomic level structure of the spliceosome just before mRNA formation.
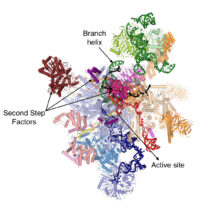
During gene expression, genetic material is transcribed into pre-messenger RNA (pre-mRNA) which contains introns, non-coding sequences that must be removed before the mature RNA (mRNA) can be used for protein synthesis. The removal of introns from pre-mRNAs is catalysed by the spliceosome, a macromolecular machine composed of both protein and RNA. The spliceosome recognizes the 5’- and 3’-ends of the intron and mediates excision of the intron in a two-step reaction, undergoing several dynamic rearrangements during the process. Kiyoshi’s group have found that before the second reaction step can occur, the helicase Prp16 remodels the spliceosome’s active site and promotes dissociation of first step protein factors, binding of second step factors and recognition of the 3’-end of the intron, thus allowing production of the mRNA.
Sebastian Fica, Chris Oubridge, Wojciech Galej and Max Wilkinson in Kiyoshi’s group, with the help of Xiaochen Bai and Andy Newman, used yeast to make an extract containing active spliceosomes which was then incubated with pre-mRNA containing a mutation at the 3’-end of the intron to prevent mRNA formation. These spliceosome-pre-mRNA complexes were purified and visualised using electron cryo-microscopy, resulting in a 3D image of the complex that allowed them to build a near-atomic model of all of the spliceosome’s RNA and protein components.
It is estimated that 15-30% of diseases are caused as a result of errors in splicing. This work provides a molecular framework for investigating how splice sites are recognized and thus for determining the mechanisms that lead to errors in splicing. Using knowledge acquired from this work, future clinical researchers will be able to target such errors more specifically and perhaps determine novel strategies to correct them.
This work was funded by the MRC, the European Research Council, EMBO, an EU Marie Sklodowska-Curie fellowship and a Rutherford Memorial Cambridge Scholarship.
Further references:
Paper in Nature
Previous Insight on Research article: Structure of the catalytic spliceosome
Previous Insight on Research article: Cryo-EM sheds new light on the spliceosome activation process
Kiyoshi’s group page