Researchers at the LMB have solved the elusive 3D structure of activated Parkin, an enzyme implicated in early-onset Parkinson’s disease. Led by David Komander’s group in the LMB’s PNAC Division, in collaboration with the LMB’s Biological Mass Spectrometry facility, this new work reveals insights into previously unstudied parts of this important protein and helps explain why families with certain mutations in Parkin suffer from early-onset Parkinson’s disease.
Whilst most cases of Parkinson’s disease arise spontaneously with no known cause, mutations in several genes are linked to an early-onset familial form of the disease. Among these are genes encoding PINK1 (an enzyme which modifies its protein substrates by adding a phosphate group – phosphorylation) and Parkin (an enzyme which attaches a small protein called ubiquitin to other protein substrates – ubiquitination).
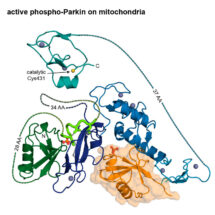
PINK1 and Parkin function together in the clearance of damaged mitochondria from cells, a process which if impaired in neurons can lead to Parkinson’s disease. Recent work by David’s group and others has generated insights into this process at the molecular level. PINK1 accumulates on damaged mitochondria and phosphorylates ubiquitin on the surface, creating a unique signal which recruits Parkin. When Parkin binds the phosphorylated-ubiquitin, its closed, inactivated structure is partially opened. This exposes Parkin’s ubiquitin-like domain which is then also phosphorylated by PINK1. Once phosphorylated, the full catalytic activity of Parkin is unleashed on the surface of the damaged mitochondria: Parkin labels the damaged organelles with ubiquitin, directing them for destruction. Until now, however, the structure of opened, active, phosphorylated Parkin remained elusive and it was unclear how phosphorylation by PINK1 could lead to full opening of Parkin and unleashing of the catalytic activity.
To reveal how the closed structure of Parkin opens for catalytic activity, PhD student Christina Gladkova, from David’s group, trapped Parkin at intermediate stages between inactive and active forms and studied their shape by mass spectrometry. Understanding how the structure changed during the transition helped her trap the active open conformation of Parkin and determine its 3D structure. Christina’s work also identified a new class of mutations which prevent Parkin activation – without Parkin activation, damaged mitochondria cannot be cleared from neurons, leading to early-onset forms of Parkinson’s disease.
Approximately 10% of Parkinson’s disease cases are due to an underlying genetic cause, commonly resulting in Parkin malfunction. The ability to distinguish between the active and inactive forms of Parkin stemming from this new work will enable the design of therapies that help prevent neuronal loss by increasing the proportion of active Parkin. Measurement of the proportion of active Parkin may give rise to diagnostic tools to assess neuronal health. In the future, accurate classification of early-onset Parkinson’s disease patient mutations could aid targeted therapy.
This work was funded by the MRC, European Research Council, Michael J. Fox Foundation and the Lister Institute for Preventative Medicine.
Video to show the transition of Parkin from its inactive to active state
Further references:
Paper in Nature
Previous Insight on Research article: New insights into ubiquitin phosphorylation and the development of early onset Parkinson’s disease