Genes are encoded in DNA and need to be copied into an intermediate mRNA molecule that contains the instructions to allow synthesis of protein. Almost every mRNA has a repetitive sequence at one end called a poly(A) tail. The length of this tail specifies the amount of time that the mRNA is present in the cell, and how often it is translated into protein. Errors or changes in the tail are found in human diseases including β-thalassemia, thrombophilia and cancer, as well as viral infections.
Now, work by Lori Passmore’s group in the LMB’s Structural Studies Division, in collaboration with Carol Robinson’s lab at the University of Oxford, has revealed new insights into how the poly(A) tail is added to the end of mRNAs. They discovered that the 14 different protein subunits are assembled via three separate modules into the molecular machine that adds the tails.
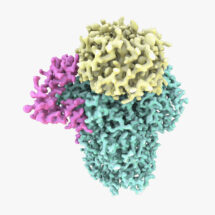
Lori’s group used electron cryo-microscopy (cryo-EM) to determine a 3.5 Å structure of one of these modules: the polymerase module. The structure reveals a striking similarity to other cellular machines that interact with DNA or RNA. The group were also able to reconstitute the activity of the complex with fully purified components to add poly(A) tails onto RNAs. These studies, in combination with the structure and mass spectrometry, show that the polymerase module acts as a hub to bring together all the factors required for specific poly(A) tail addition.
These findings have allowed us to visualise details of cellular machines that are fundamental to life. These machines are found in every eukaryotic cell and play a role in controlling expression of genes into protein. They also reveal new insight (building on previous crystal structures from other groups) into how viruses, such as influenza, are able to highjack the host cell machinery.
This work was funded by the MRC, the European Research Council (ERC), EMBO, Gates Cambridge and used electron microscope facilities at MRC LMB and Diamond Light Source.
Further references:
Paper in Science
Lori’s group page
Carol Robinson’s group page