Cryo-electron tomography reveals how the protein GOLPH3 enables selective retention of key Golgi enzymes amid constant cargo flow
The Golgi apparatus is a crucial sorting structure, operating akin to a post office within the cell, receiving, modifying and directing proteins to their correct locations. The apparatus itself is a stack of membrane-bound compartments called cisternae, each with a defined protein content critical for its function. So why don’t these proteins get swept up in the constant flow of cargo proteins that move through the Golgi stack? Sean Munro’s group, in the LMB’s Cell Biology Division, have worked with John Briggs’ group, formerly in the LMB’s Structural Studies Division and now at the Max Planck Institute of Biochemistry in Munich, to investigate just this. Together, they have found how the adaptor protein GOLPH3 enables vesicles to selectively retain a key set of proteins within the correct part of the Golgi.
Newly made secretory and membrane proteins are transported from the endoplasmic reticulum (ER) in small membrane-bound vesicular carriers. These vesicles deliver the proteins to the first cisternae of the Golgi stack and they then move through the stack before leaving to other locations in the cell. One model for how they move through the stack is by the cisternae maturing or progressing like a conveyor belt whilst vesicles recycle the Golgi resident proteins to their correct location. The vesicles that bud from the Golgi are formed by coat proteins which select specific cargoes and chaperone them to specific locations. Of these, COPI-coated vesicles are responsible for retrieving escaped ER proteins back to the ER and recycling Golgi resident proteins within the stack. This relies on COPI correctly distinguishing these cargoes amongst a vast variety of proteins traversing the Golgi apparatus.
To better understand this, Sean and John’s groups focused on GOLPH3, a protein which binds to the cytoplasmic tails of many Golgi enzymes and links them to the COPI coat for retrieval. The team reconstituted GOLPH3-containing COPI-coated vesicles in vitro and used cryo-electron tomography (cryo-ET) to reveal its structure. This was a significant undertaking and required manual identification of over 4,800 vesicles and buds.
This work revealed that GOLPH3 interacts with COPI at two distinct sites: its N-terminal domain binds to COPI’s β-COP subunit and its C-terminal domain binds to the β-propeller of the α-COP subunit. This latter connection obstructs the main binding site for ER retrieval signals. The lipid PI4P, which is abundant only in the later cisternae of the Golgi apparatus is a major membrane component recognised by GOLPH3. By binding to both COPI and PI4P, GOLPH3 creates a “coincidence detection” mechanism, which ensures that it is only present in COPI vesicles later in the stack and so prevents interference with ER retrieval in the earlier cisternae.
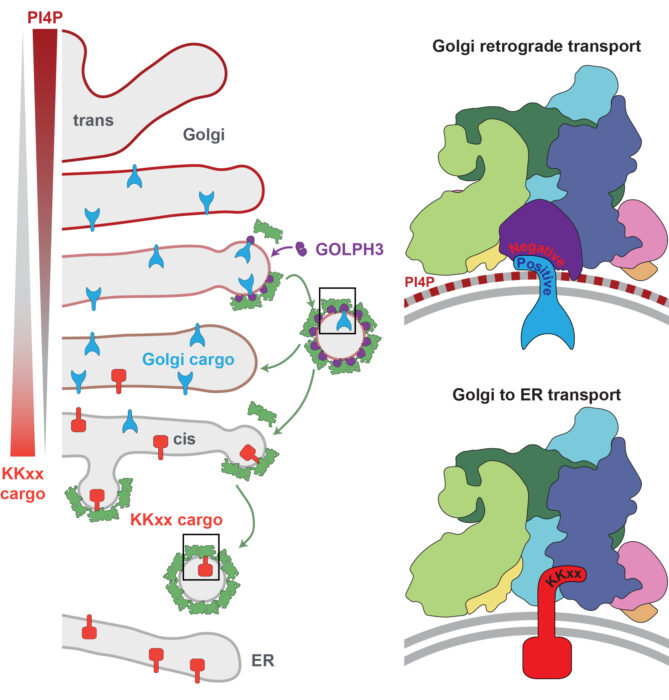
Finally, the team performed site-directed mutagenesis, altering specific amino acids in GOLPH3 to understand how it recognises different Golgi enzymes. They found that mutations disrupting the α-COP binding site totally revoked GOLPH3’s ability to retain Golgi enzymes. Additionally, they found that different regions of GOLPH3’s membrane-facing surface bind different cargo proteins. The presence of multiple, cargo-specific binding sites on GOLPH3 explains why prior research was unable to identify a single retention signal for Golgi enzymes.
These findings support the cisternal maturation model of Golgi trafficking, in which each cisterna matures over time, retaining its cargo, whilst Golgi resident enzymes are recycled backwards by COPI-coated vesicles to earlier cisternae. This work illustrates how GOLPH3 enables COPI to switch roles during cisternal maturation: retrieving ER proteins in early compartments and recycling Golgi enzymes in later ones. This challenges a previously proposed model of Golgi trafficking, the vesicular transport model, in which cisternae and Golgi enzymes were thought the be static and cargo moves between them via vesicles.
This research also holds wider implications for human health, as GOLPH3 is often amplified in certain cancers and is involved in glycosylation, a process critical for immune recognition and cell signalling. Further studies to better understand the function of GOLPH3 could therefore prove vital to new therapeutic interventions.
This work was funded by UKRI MRC, the Max Planck Society, the Wellcome Trust and EMBO.
Further references
The mechanistic basis of cargo selection during Golgi maturation. Taylor, R. J., Zubkov, N., Ciazynska, K. A., Kaufman, J. G. G., Tagiltsev, G., Owen, D. J., Briggs, J. A. G. and Munro, S. Science Advances
Sean’s group page
John’s group page at the Max Planck Institute for Biochemistry
Previous Insight on Research articles
A database to identify important genes of unknown function for analysis
Tail of SARS-CoV-2 Spike protein is optimised to reach the cell surface causing infection to spread to neighbouring cells