 Ribosomes are cellular molecular machines that link amino acids together in the order specified by messenger RNA (mRNA) to make proteins. Near the end of the mRNA molecule a specific nucleotide sequence, known as a stop codon, signals for protein synthesis to terminate by recruiting release factors that release the newly made protein from the ribosome and recycle the ribosome to start another round of protein synthesis.
Ribosomes are cellular molecular machines that link amino acids together in the order specified by messenger RNA (mRNA) to make proteins. Near the end of the mRNA molecule a specific nucleotide sequence, known as a stop codon, signals for protein synthesis to terminate by recruiting release factors that release the newly made protein from the ribosome and recycle the ribosome to start another round of protein synthesis.
However, some mRNA molecules are defective. Problems in making mRNA, or enzymes in the cell that cut mRNA, can result in mRNA molecules that lack stop codons. Ribosomes are unable to detect these corrupt mRNAs before they begin synthesizing proteins from them, but without a stop codon release factors are not recruited and the ribosomes stall at the end of the mRNA. Over time, the accumulation of stalled ribosomes is lethal to the cell.
This is a problem in all kingdoms of life. However, bacteria and eukaryotes (including mammals) have evolved entirely separate mechanisms to relieve stalled ribosomes. Now researchers in the LMB’s Structural Studies and Cell Biology divisions have used electron cryomicroscopy (cryo-EM) to reveal new insights into these mechanisms.
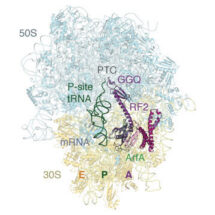
In a paper published in Science, lead author Nathan James, together with Alan Brown and Yuliya Gordiyenko in Venki Ramakrishnan’s group, describe how a small protein known as ArfA can recognize stalled ribosomes in bacteria by binding within the mRNA entry channel that in normal circumstances would contain mRNA. Once bound to the ribosome, it acts like an mRNA stop codon and recruits a release factor to the ribosome. Recruitment of the release factor allows normal termination to occur and the ribosome to be recycled.
Conformational changes of ArfA and RF2
In eukaryotes, an unrelated protein, Pelota, recognizes stalled ribosomes. In a paper in Cell, Susan Shao from Ramanujan Hegde’s group together with co-lead authors Jason Murray and Alan Brown from Venki Ramakrishnan’s group, show how this protein structurally resembles the eukaryotic release factor but with a long loop that can reach down into the mRNA entry channel and recognize stalled ribosomes. After recognition, Pelota activates a pathway that relieves the stalled ribosomes.
It is hoped that these new insights will enrich our understanding of the fundamental process of protein synthesis and also help to develop new antibiotics that target ribosomal processes in pathogenic bacteria while leaving human ribosomes unaffected.
This work was funded by the MRC, the Wellcome Trust and St John’s College, Cambridge.
Further references:
Paper in Science
Paper in Cell
Venki’s group page
Ramanujan’s group page