Structure of the cancer-associated adhesion GPCR ADGRL4 and a G protein offers a structural blueprint for the development of tumour-targeting therapies
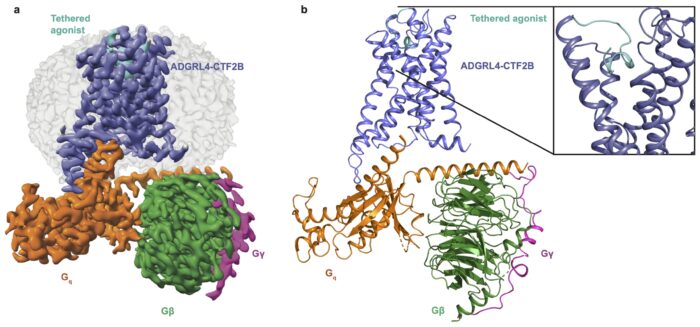
G protein-coupled receptors (GPCRs) are a vast family of proteins found on the surface of cells, responsible for detecting external signals and relaying these inside cells to trigger diverse responses. Thirty-two of these GPCRs can be defined as adhesion GPCRs (aGPCRs), which are vital to cell adhesion, migration and tissue development. They also have key roles in immune function and tumour development, making them a potential key target for therapeutics. One of these, ADGRL4, is notably upregulated within the tumour microenvironment and implicated in tumour pathogenesis in a wide range of aggressive cancers. Previous research has suggested that ADGRL4 does not signal through GPCR pathways. However, a new study led by David Favara, postdoc in Chris Tate’s group in the LMB’s Structural Studies Division and Clinical Lecturer in Medical Oncology at the Department of Oncology, University of Cambridge and Cambridge University Hospitals, has found that ADGRL4 does in fact couple weakly to a G protein.
Using highly sensitive bioluminescent assays, the group found that ADGRL4 interacts with the G protein Gq, though much less strongly than is typical for GPCR interactions. No significant interaction was found with other G proteins. To visualise ADGRL4 in its active state bound to Gq, the team used electron cryomicroscopy (cryo-EM) to determine its structure. The resulting 3.1 Å resolution structure revealed that the receptor’s overall shape matches other aGPCRs, but at its Gq coupling it has fewer contact points than usual, thus explaining its weak interaction and how its association was previously missed.
The cryo-EM structure also revealed the activation mechanism of ADGRL4; it is switched on by its own ‘tethered agonist’, a hidden peptide revealed when part of the receptor is removed. This peptide binds inside a pocket of the receptor and triggers a structural change to allow Gq binding to occur. Mutagenesis studies targeting eight different amino acid residues contacting the receptor pocket found that the residues Met412 and Phe409 are most critical for activation as well as identifying three conserved motifs which, when mutated, significantly reduce or abolish Gq coupling.
This study provides new insight into how ADGRL4 functions, overturning the previous assumption that it didn’t signal through G proteins. By capturing the first high-resolution structure of ADGRL4 in its active state, the group have gained a detailed structural blueprint of its activation mechanism. This knowledge is a critical step forward to designing new drugs which specifically block ADGRL4, a key target owing to its association with aggressive cancers. This could lead to new therapeutic interventions for cancers which are currently difficult to treat, such as glioblastoma and other metastatic tumours associated with elevated ADGRL4 production. Beyond cancers, the study holds wider implications by advancing understanding of adhesion GPCR biology, which opens new doors to create targeted treatments in other diseases which rely on these receptors.
In recognition of the significance of this work to the field of oncology, David Favara has been announced as the 2025 recipient of the McElwain Prize from the Association of Cancer Physicians/UK Society for Medical Oncology.
This work was funded by UKRI MRC, Cancer Research UK and the Department of Oncology, University of Cambridge.
Further references
Structure of the Gq-coupled adhesion receptor ADGRL4, a GPCR implicated in cancer. Chen, Q., Gusach, A., Diamante, A., Patel, J. C., Edwards, P. C., Tate, C. G. and Favara, D. M. Nature Communications
Chris’ group page
Previous Insight on Research articles
Structural study reveals a novel activation mechanism for the fungal GPCR, Ste 2
First structure of a fungal GPCR
High-resolution structure of a GPCR-arrestin complex