Uncovering the structure of M1 matrix proteins, and their arrangement within the influenza A virus, suggests mechanisms for two critical processes in the infection cycle
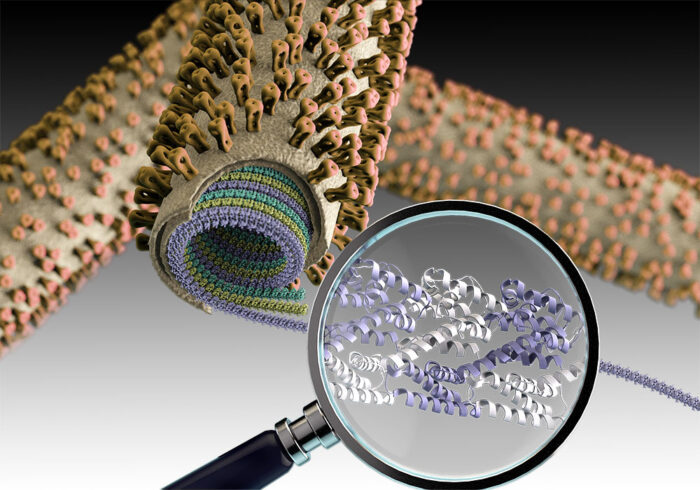
The influenza A virus is surrounded by a lipid bilayer that forms the outermost layer much like the plasma membranes on our own cells. Immediately under this lipid bilayer is a dense protein layer formed from M1 matrix protein. It has been thought that formation of this protein layer plays a crucial role in virus assembly, but how it might do this has remained unanswered. John Briggs’ group, in the LMB’s Structural Studies Division, has now uncovered the detailed structure and arrangement of M1 proteins within a virus particle, providing new clues to how the influenza virus is assembled and disassembled.
M1 is a small, two-domain protein and the most abundant protein found in the influenza A virus. The structure of one of its two domains, known as the N-terminal domain, has been previously resolved by crystallography, but this was not sufficient to show how this protein might mediate virus assembly. Interestingly, influenza A virions can adopt spherical or filamentous morphologies, so an assembly mechanism needs to allow these different possible shapes.
Members of John’s group, Julia Peukes, Xiaoli Xiong, Simon Erlendsson, and Kun Qu, used electron cryo-microscopy (cryo-EM) and electron cryo-tomography (cryo-ET) to provide the first description of the full-length structure of M1 and see how it is arranged within virus particles. To do this, Julia infected mammalian cells with influenza A virus, leading to production of new viruses. She then captured highly magnified images of the infected cells from which new virions were growing and obtained 3D reconstructions of the virions and structures of individual matrix proteins within them.
Xiaoli purified the matrix protein in order to characterise its structure and dynamics using NMR with support from Simon and Stefan Freund in the LMB’s NMR facility. Xiaoli also identified a condition under which multiple molecules of the protein form into an ordered helical assembly, which he imaged with Kun using cryo-EM to obtain a high-resolution structure.
Analysis of the structures suggests ways in which the virus assembles and disassembles. The team discovered that M1 forms multiple, parallel, linear strands arranged in a helical fashion. The previously unresolved structure of the second of M1’s two domains, the C-terminal domain, was seen to fold to interact with the next M1 molecule in the chain, providing insight into how viral assembly via growth of strands of M1 might take place. Furthermore, their detailed structures also showed a cluster of 5 histidine residues coming from three successive M1 monomers. The team suggest this cluster could function as a pH-sensitive switch providing a mechanism for virus disassembly during entry into a cell.
Influenza causes very large numbers of cases of severe disease and death every year and new flu strains can cause global pandemics. Understanding the role of the matrix protein M1 in assembly and disassembly during virus entry, two critical processes in the infection cycle, may help to target M1 polymerisation or depolymerisation and interfere with virus infection.
The work was funded by UKRI MRC, ERC, EMBL, and the Deutsche Forschungsgemeinschaft, and was performed in collaboration with scientists at the University of Heidelberg and the Francis Crick Institute.
Further references
The native structure of the assembled matrix protein 1 of influenza A virus. Peukes, J., Xiong, X., Erlendsson, S., Qu, K., Wan, W., Calder, LJ., Schraidt, O., Kummer, S., Freund, SMV., Kräusslich, HG., Briggs, JAG. Nature doi:10.1038/s41586-020-2696-8.
John’s group page