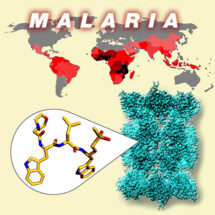
 Every year hundreds of millions of people worldwide are affected by Malaria and nearly half a million die from the disease. More than two thirds of those dying are children under five. The disease is caused by parasites passed to humans through the bites of infected mosquitoes, with Plasmodium falciparum being the parasite responsible for the most severe form of malaria. Increasingly, the parasites are becoming resistant to existing anti-malaria treatments, leading to a major threat to human health and the urgent need to develop new anti-malaria drugs. An important step forward in this quest has been made by a collaboration of research groups: Matthew Bogyo’s group at Stanford University School of Medicine, Paula da Fonseca’s group in the LMB’s Structural Studies Division, and Leann Tilley’s group at the University of Melbourne, who have determined, at the molecular level, the structure and function of the Plasmodium falciparum proteasome and used this information to design a new compound with promising anti-malarial properties.
Every year hundreds of millions of people worldwide are affected by Malaria and nearly half a million die from the disease. More than two thirds of those dying are children under five. The disease is caused by parasites passed to humans through the bites of infected mosquitoes, with Plasmodium falciparum being the parasite responsible for the most severe form of malaria. Increasingly, the parasites are becoming resistant to existing anti-malaria treatments, leading to a major threat to human health and the urgent need to develop new anti-malaria drugs. An important step forward in this quest has been made by a collaboration of research groups: Matthew Bogyo’s group at Stanford University School of Medicine, Paula da Fonseca’s group in the LMB’s Structural Studies Division, and Leann Tilley’s group at the University of Melbourne, who have determined, at the molecular level, the structure and function of the Plasmodium falciparum proteasome and used this information to design a new compound with promising anti-malarial properties.
The proteasome is a protein recycling complex responsible for the breakdown and removal of old, damaged or unnecessary proteins from cells. Its proper function is critical in the regulation of key mechanisms such as cell division and the controlled removal of damaged cells from the organism. Matthew Bogyo’s group have previously shown that drugs that block the proteasome from degrading proteins are toxic to the Plasmodium falciparum parasite. However, all human cells are also dependent on proper proteasome function. Therefore, it is important to determine, at the molecular level, the differences between the human and the Plasmodium proteasomes in order to develop compounds that selectively stop the parasite proteasome functioning, while leaving the human complex unharmed.
At the LMB, Paula’s group looked at the Plasmodium falciparum proteasome in the electron microscope at ultra-low temperatures (cryo-EM). Following computational processing of the images, they displayed the parasite proteasome complex in three-dimensions with great detail, down to a resolution of around 3.6 Angstroms (or 3.6 ten-billionths of a metre), including the details of a prototype anti-malarial drug blocking one of its active sites. By carefully comparing the Plasmodium falciparum and the human proteasomes they could understand why the new drugs, developed in Matthew Bogyo’s lab, can specifically block the parasite proteasome while evading binding to the human complex. This information can now be exploited to design more effective compounds suitable for therapeutic use.
The authors have demonstrated the potential of the new compounds that can block the function of the Plasmodium falciparum proteasome, while avoiding interaction with the human complex, to be developed as next generation anti-malaria drugs. With the increase of resistance to current anti-malaria drugs, the development of new, efficient, anti-malarial treatments is most urgent.
The work was funded by NIH, MRC, Agency for Science, Technology and Research (A*STAR) Singapore, Netherlands Organisation for Scientific Research, Program for Breakthrough Biomedical Research, Sandler Foundation, American Heart Association, Australian Research Council and the Australian National Health and Medical Research Council.
Further references:
Paper in Nature
Paula’s group page
Matthew Bogyo’s group page
Leann Tilley’s group page
Stanford University Press Release
C&E News Article