Structures of active Rag dimers bound to mTORC1 provide greater understanding of control of cell growth and identify potential targets for development of new cancer drugs
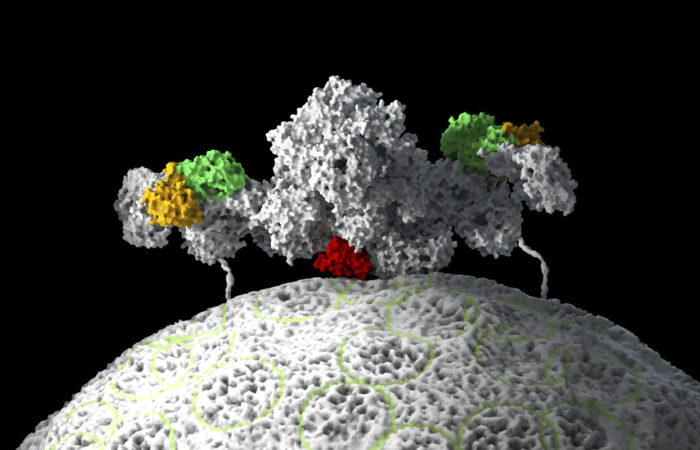
Control of cell division is crucially important, as unregulated cell division is a hallmark of cancer. mTORC1 protein kinase is an ancient enzyme complex and master regulator of growth and metabolism that integrates signals relating to nutrient availability, energy, and growth factors. Activation of mTORC1 is driven by proteins called Rags that sense nutrient abundance. However, some cancer-causing mutations make Rag proteins active even in the absence of nutrients and drive cells to divide when they shouldn’t. Roger Williams’ group, in the LMB’s PNAC Division, has now shown how active Rags bind to mTORC1 and how a cancer-associated mutation can make a Rag protein continuously active.
When nutrients are lacking, mTORC1 is inactive and distributed in the cytoplasm, but when nutrients become abundant, mTORC1 moves quickly to the surface of lysosomes, where it is activated by RHEB GTPase. This movement is controlled by Rag proteins that are tethered to the surface of the lysosome through a docking station protein. When activated by nutrient abundance, Rag proteins bind to mTORC1 as a pair or dimer: one of the pair is bound to GTP (guanosine triphosphate), while the other is bound to GDP (guanosine diphosphate). However, the precise way in which Rags transition between active and inactive forms, and how active forms bind to mTORC1, was not known.
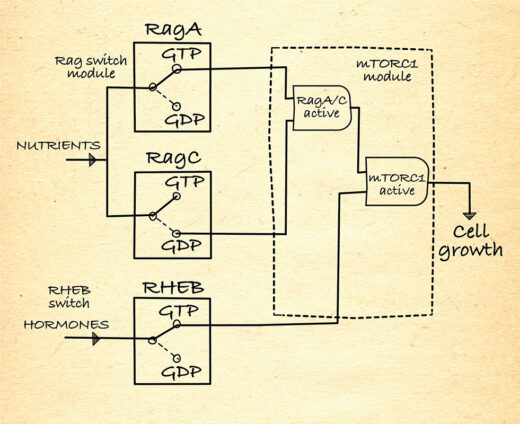
How is mTORC1 regulated?
As depicted in this AND-gate diagram, mTORC1 is only active when the Rags are activated by nutrient abundance, so that mTORC1 is recruited to the lysosomes, and RHEB is activated by hormones. This ensures cell growth only occurs when it is needed and nutrients are plentiful.
Madhan Anandapadamanaban and other members of Roger’s group, in collaboration with David Sabatini’s group at the Whitehead Institute in the USA, have now solved structures of Rag proteins in complex with mTORC1. By using electron cryo-microscopy, X-ray crystallography, and Hydrogen-Deuterium Exchange Mass Spectrometry, Roger’s group has been able to develop our understanding of the mechanism of Rag function in activation of mTORC1. In particular, the team showed how cancer-causing mutations in one Rag protein, RagC, result in the loss of structural elements in a region that functions as a switch, thereby fixing it permanently in an active state.
By determining these structures, Madhan also demonstrated why only the active forms of Rag proteins bind to mTORC1 and importantly that this binding doesn’t immediately activate mTORC1. This allowed the team to propose the organisation of the entire system in which activated Rag proteins bind mTORC1 and recruit it to the lysosome surface where it is activated by another protein.
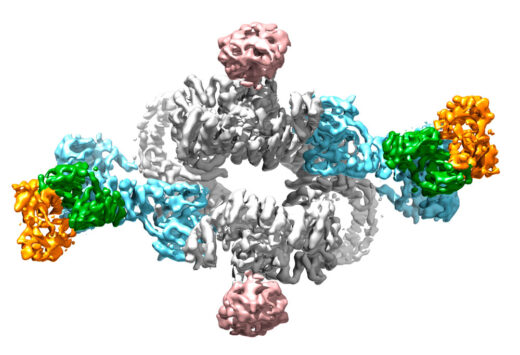
By providing a structural understanding of this process and the demonstration of how cancer can cause overactivation of it, this work could provide the basis for development of new targeted drugs against these proteins that could be used to fight cancer. Rapamycin is an mTORC1 inhibitor that is used widely for various clinical applications, particularly because of its immunosuppressant function, but it has limitations and side effects. More targeted drugs against this signalling pathway could be of great benefit to many patients.
The work was funded by the MRC, CRUK, FEBS, EMBO, St Catherine’s College, Tuberous Sclerosis Association, Royal Society of Chemistry, Company of Biologists, NIH, the US Department of Defense, and Lustgarten Foundation.
Further references
Architecture of human Rag GTPase heterodimers and their complex with mTORC1. Anandapadamanaban, M., Masson, GR., Perisic, O., Berndt, A., Kaufman, J., Johnson, CM., Santhanam, B., Rogala, KB., Sabatini, DM., Williams, RL. Science
Roger’s group page
David Sabatini’s group page
Previous Insight on Research: Insight into the complex 3D topology of the TOR enzyme