Electron cryo-tomography and sub-tomogram averaging used to determine structure of native γTuRC capping microtubules from enriched Spindle Pole Bodies
Microtubules, which are comprised of abundant tubulin polymers, support cell life in numerous ways, including underpinning cellular architecture and forming the mitotic spindle structures that mediate DNA segregation during cell division. Microtubules are formed by the process of nucleation, orchestrated by a large protein complex named the γ-Tubulin Ring Complex (γTuRC). The molecular mechanisms behind this nucleation have, so far, remained unclear. To tackle this, David Barford’s group, in the LMB’s Structural Studies Division, in collaboration with John Kilmartin, Emeritus Scientist in the LMB’s Cell Biology Division, extracted the budding yeast spindle pole body (SPB), the microtubule organising centre in yeast, and used electron cryo-tomography (cryo-ET) and sub-tomogram averaging to determine the structure of its native γTuRC capping microtubules.
The project, led by postdoc Tom Dendooven and Ph.D. student Stanislau Yatskevich, revealed that the γTuRC adopts the microtubule helical geometry to template microtubule nucleation. In addition, the group discovered a novel coiled-coil protein factor, and suggest that this could catalyse the initial steps of microtubule nucleation.
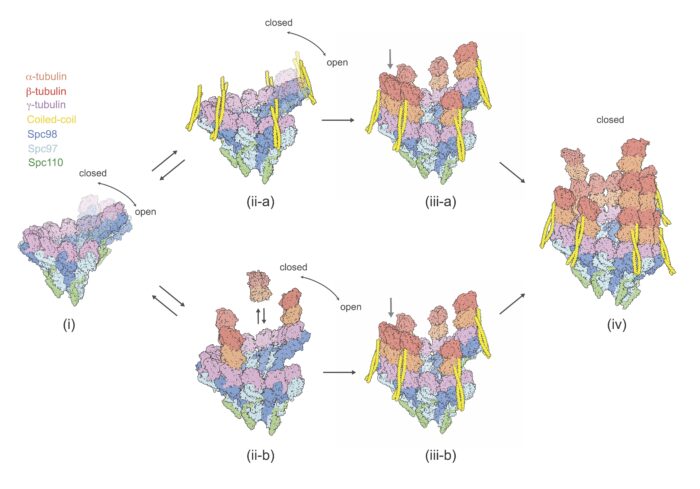
To enrich the SPBs, Yeast nuclei were prepared and solubilised after which SPBs were isolated by sucrose gradient centrifugation. Following this, Tom and Stanislau froze the SPBs on grids and performed cryo-ET. To further investigate the identity of the coiled-coil proteins, the group worked with Juri Rappsilber’s group at Technische Universität, Berlin to conduct further analysis using cross-linking mass spectrometry.
This structural study provides unprecedented insight into the molecular pathway behind microtubule nucleation. Beyond its importance in advancing our understanding of core cellular processes, the work also holds potential clinical implications, as proteins responsible for the formation and ongoing function of the mitotic spindle can be used as targets by anti-cancer drugs seeking to halt cell division, and hence cancer progression. An improved knowledge of the process of spindle formation at a molecular level will help to identify new targets for such drugs.
This work was funded by UKRI MRC, Cancer Research UK, Boehringer Ingleheim Fonds, the Wellcome Trust and Deutsche Forschungsgemeinschaft (German Research Foundation).
Further references
Structure of the native γ-tubulin ring complex capping spindle microtubules. Dendooven, T., Yatskevich, S., Burt, A., Chen, ZA., Bellini, D., Rappsilber, J., Kilmartin, JV., Barford, D. Nature Structural & Molecular Biology
David’s group page
John Kilmartin’s page
Juri Rappsilber’s group
Previous Insight on Research articles
Molecular mechanisms of mitotic error correction revealed by cryo-EM
Human inner kinetochore structure reveals mechanism for binding DNA during mitosis
How chromosomes are bound to be separated in cell division