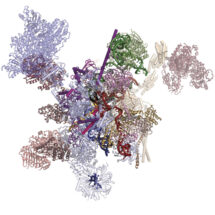
Although humans have a similar number of genes as flies, part of our greater complexity comes from a process called alternative splicing, in which multiple different variants of proteins can be made from a single gene. This process is controlled by a molecular machine called the spliceosome. Until recently, much of the work on spliceosomes has been done using yeast spliceosomes as this system is well conserved and works very similarly across all eukaryotes. However, very little alternative splicing happens in yeast, so structures of human spliceosomes are needed for understanding of how this is controlled. Kiyoshi Nagai’s group in the LMB’s Structural Studies Division has now revealed the first structure of a human spliceosome frozen in a position immediately after the reaction has finished.
When the DNA code is read in order to produce protein, it is first transcribed into RNA, before that RNA is translated into a protein. However, initially the RNA is in the form of pre-messenger RNA (pre-mRNA), which has to be processed into mature mRNA by the spliceosome. This is because the pre-mRNA is composed of two types of regions: exons and introns. Only exons code for the protein and therefore introns must be removed so that the exons can be joined together to produce one continuous protein coding chain. This process is called splicing and is made more complex by the possibility of alternative splicing, in which different mature mRNAs can be produced by joining multiple exons in different combinations. Studying the structure of the spliceosome allows a greater understanding of how this choice is made.
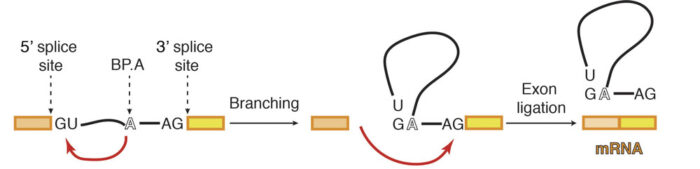
Previous work from Kiyoshi’s group provided a structural explanation for the process of splicing in two steps: first, a cut is made at the end of one exon and second, that end is joined to the start of the next exon. This second step is termed exon ligation. Once exon ligation occurs, a protein called Prp22 allows release of the mature mRNA. Using a mutant form of Prp22 to stall the spliceosome at the point just after exon ligation, Sebastian Fica and Chris Oubridge, with the help of Max Wilkinson and Andy Newman, all from Kiyoshi’s group, have used cryo-EM to reveal the first structure of a post-reaction human spliceosome and identified four proteins that had never been seen in human spliceosomes before.
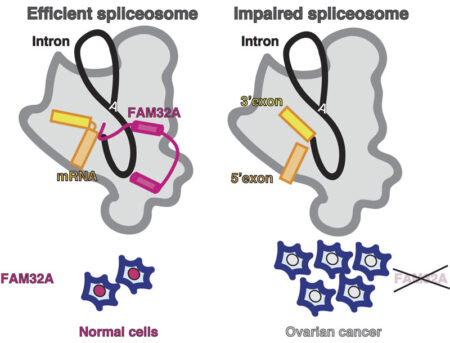
One of these proteins, FAM32A, was seen to bind to the first exon and promote the binding of the second exon at the active site. Additionally, Sebastian showed that this protein was required for efficient exon ligation to occur. This is the first multicellular animal-specific protein to be found in the active site of the spliceosome. This structure provides a framework for understanding the role of mammalian-specific exon ligation proteins, as it is thought that there will be other proteins that cooperate with FAM32A, depending on the cell-type and the mRNA to be spliced.
It is estimated that 15-30% of hereditary diseases are due to errors in splicing, so understanding how this process is controlled in the human spliceosome is very important. Interestingly, FAM32A has previously been found to promote formation of mature mRNAs for genes that direct cellular suicide. Its loss can then allow inappropriate survival of cells and indeed, reduced production of FAM32A is associated with ovarian cancer, which also makes this the first direct functional link between splicing catalysis and cancer.
This work was funded by the MRC, ERC, EMBO, Marie Sklodowska-Curie Actions, and a Cambridge-Rutherford Memorial PhD Scholarship.
Further references:
A human post-catalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Fica, SM., Oubridge, C., Wilkinson, ME., Newman, AJ., Nagai, K. Science 363(6428): 710-714
Kiyoshi Nagai’s group page
Previous Insight on Research: Spliceosome catalysis: the completed puzzle
Downloadable spliceosome structures from Kiyoshi’s group