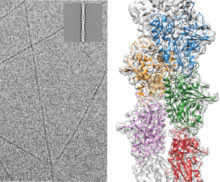
 The structure of the actin-like protein ParM in different states has been revealed by Tanmay Bharat and Jan Löwe in collaboration with Garib Murshudov from the LMB and Carsten Sachse from the EMBL. Using electron cryo-microscopy (cryo-EM) they determined the structure of ParM to almost atomic resolution and revealed how this protein carries out the process of plasmid DNA segregation in growing bacterial cells.
The structure of the actin-like protein ParM in different states has been revealed by Tanmay Bharat and Jan Löwe in collaboration with Garib Murshudov from the LMB and Carsten Sachse from the EMBL. Using electron cryo-microscopy (cryo-EM) they determined the structure of ParM to almost atomic resolution and revealed how this protein carries out the process of plasmid DNA segregation in growing bacterial cells.
Plasmids are small circular pieces of DNA, most commonly found inside bacterial cells, which confer an advantage to the cell. Before the bacterial cell divides, copies of the plasmid need to move apart so that each daughter cell contains an equal number of copies. For some plasmids, the movement takes place on a spindle made up of filaments of the ParM protein. These ParM filaments bind to a ParR protein, which then binds to the plasmid, forming a ParMRC complex. As further subunits of ParM are added, the two copies of the plasmid are moved to opposite sides of the dividing cell.
Several models have been proposed for how this process could happen. To investigate these, ParM filaments were imaged using whole-cell electron cryo-tomography to view the filaments in their native environment. This revealed for the first time the details of how ParM filaments bind together to form antiparallel doublets, with the plasmid-binding end facing opposite sides of the cell. These results explain how sister plasmids could be pushed apart as the filaments grow, and have led to a model for plasmid segregation uncoupled with bacterial cell division and with a separate spindle for each pair of sister plasmids. This study also showcases the range of information that cryo-EM can deliver for structural and cell biology research from high-resolution structures to molecular resolution contextual information from whole-cell tomography.
This research could provide important clues to help combat the widespread problem of antibiotic resistance mediated by plasmids. Many bacteria use the ParMRC system to maintain important genes, such as those responsible for antibiotic resistance. This research tackles the important problem of bacterial DNA segregation and how plasmids are maintained in bacterial populations.
This research was funded by the MRC, FEBS, the Wellcome trust and EMBO.