Total synthesis of E. coli with just 57 codons is a radical rewrite of the genetic code, proving life’s capabilities with seven fewer codons

The genetic code, which has been nearly universal across all known life for billions of years, utilises 64 triplets of DNA and RNA bases, known as codons, to encode 20 amino acids and stop signals, which together are used to build proteins. However, this is not an optimised system as several codons contain duplicate instructions for the same few amino acids or stop signals. Jason Chin’s group in the LMB’s PNAC Division previously designed and synthesised an entire E. coli genome with just 61 codons, named Syn61, showing that it is possible for life to function with a simplified genetic code. The group have now gone a step further and removed seven codons to synthesise Syn57, a strain of E. coli which utilises just 57 codons.
Specifically, in a monumental undertaking led by Wes Robertson, Fabian Rehm, Martin Spinck, Raffael Schumann and Rongzhen Tian, the group eliminated four of the six codons which encode the amino acid serine, two of the four codons for the amino acid alanine and one stop codon. Each of these were replaced by synonymous codon substitutes, capable of producing the same end products. In total, this required recoding over 101,000 codons across a 4 Mb synthetic genome.
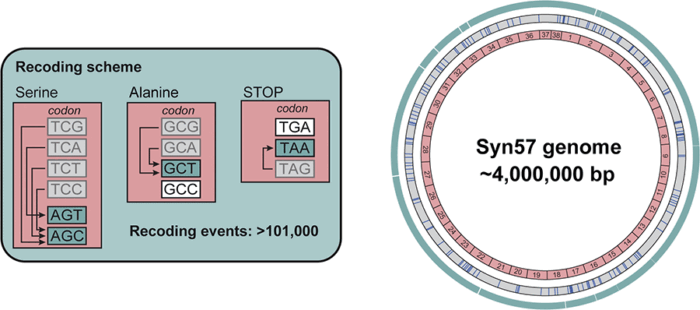
To manage this sizeable task, the genome was divided into 38 fragments, each around 100 kb. These were individually synthesised using homologous recombination in yeast. These were then inserted into E. coli using uREXER, a variant of the Replicon Excision Enhanced Recombination (REXER) method the group previously developed which combines CRISPR-Cas9 and lambda-red recombination to replace genomic DNA with synthetic DNA in a single step.
This process also flagged problematic genome regions which resisted recoding or where the growth rate was stunted. The team addressed these using several approaches including altering the N-terminal coding sequences to optimise gene expression, refactoring overlapping genes to spur independent recoding and tweaking the codon swaps used. Once each fragment was complete, the team performed iterative steps of uREXER to stitch them together into a series of semi-synthetic strains. While mapping and fixing problematic synthetic sequences at each step, the team used a bacterial mating approach to convergently assemble the semi-synthetic strains into a final, fully synthetic strain.
The result, Syn57, is the most significantly recoded organism to date, demonstrating that life can function with a considerable compressed genetic code. Jason’s group have previously illustrated how their Syn61 strain can be reprogrammed to introduce non-canonical amino acids to synthetise entirely novel polymers. By knocking out four further codons, Syn57 has more space to introduce further non-canonical amino acids, presenting greater opportunities to expand the genetic code even further. This will allow researchers to develop innovative synthetic polymers and macrocycles. Additionally, like Syn61, future work with Syn57 will be carried out to demonstrate how it cannot read the canonical genetic code and can therefore be rendered resistant to viral infection. This could make drug manufacturing cheaper and more reliable by eliminating the costly threat that viruses currently pose to this process.
This work was funded by UKRI MRC, the European Research Council (ERC), the Wellcome Trust, the German Research Foundation (DFG), UKRI Marie Skłodowska-Curie Actions, the Boehringer Ingelheim Fonds, the Cambridge Commonwealth, European and International Trust and the Harding Distinguished Postgraduate Scholars Programme.
Further references
Escherichia coli with a 57-codon genetic code. Robertson, W.E., Rehm, F.B.H., Spinck, M., Schumann, R.L., Tian, R., Liu, W., Gu, Y., Kleefeldt, A.A., Day, C.F., Liu, K.C., Christova, Y., Zürcher, J.F., Böge, F.L., Birnbaum, J., van Bijsterveldt, L. and Chin, J.W. Science
Jason’s group page
Wes’s group page
Previous Insight on Research articles
Refactoring the genetic code to create organisms protected by a genetic firewall
Cells reprogrammed for genetically encoded polymer synthesis and viral resistance
Creating an entire bacterial genome with a compressed genetic code