A crystal structure reveals the precise arrangement in which a TRIM21 dimer transfers ubiquitin onto a third TRIM21 when creating the signal to recruit the proteasome
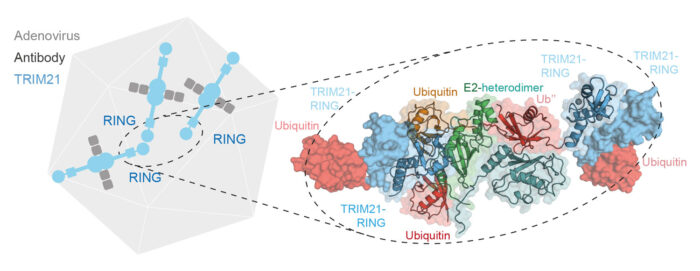
When viruses enter our cells bound to an antibody, they can be detected by the intracellular antibody receptor TRIM21. TRIM21 creates a signal, in the form of a polyubiquitin chain, that recruits the cell’s disposal system and degrades the virus. Most enzymes that add a ubiquitin modification to other proteins have a defined target, so understanding how TRIM21 can degrade any target is an important and fundamental question. David Neuhaus’ group, with Leo James’ group, has uncovered that TRIM21 itself is both the substrate and enzyme driving the ubiquitination reaction.
About 10 years ago, Leo’s group discovered that antibodies bound to viruses entering our cells trigger a response led by TRIM21 that leads to degradation of the invading virus by the proteasome. This suggested to them the possibility to exploit this capacity of TRIM21 to degrade any antibody-bound protein in the development of a technology called Trim-Away that could be used to study protein function by selectively removing a protein of interest or to treat disease by breaking down disease-causing proteins. A fuller understanding of TRIM21 will enable further enhancement of Trim-Away.
What is ubiquitination?
Ubiquitin is a small protein that can be attached to other proteins through a process called ubiquitination, which can mark proteins for degradation or regulate activity in a range of other ways. Ubiquitination involves three main steps that are driven by three different groups of enzymes, known as E1s, E2s, and E3s. TRIM21 is an E3 ubiquitin ligase enzyme that controls the final step; transfer of ubiquitin from the E2 ubiquitin-conjugating enzyme to the target protein.
To investigate how TRIM21 performs ubiquitination, Leo Kiss, a PhD student in David’s group, solved the crystal structure of TRIM21 in complex with its ubiquitin-carrying partner E2 enzyme. This structure showed a specific arrangement in which two TRIM21 molecules work together to form the active enzyme and transfer ubiquitin to a third TRIM21. Leo was also able to confirm this interesting finding of a protein serving as both the enzyme and the substrate with biochemical experiments.
Leo then wondered how long a TRIM21-anchored ubiquitin chain would have to be before the dimeric enzyme could elongate a self-bound chain rather than on a third TRIM21 molecule. With structural models and biochemical assays, the team found that after four ubiquitin units are connected to TRIM21, the chain is long enough for self-elongation. Finally, Leo could show that this enzymatic mechanism is indeed required for TRIM21 to target proteins for degradation inside human cells.
Many diseases are caused by misfunctioning proteins, so a better understanding of how TRIM21 could be used to target specific proteins for degradation could enable development of therapeutic molecules that allow Trim-Away to be used to treat a wide range of diseases by driving removal of those proteins.
The work was funded by UKRI MRC, Boehringer Ingelheim Fonds, and Wellcome Trust.
Further references
RING domains act as both substrate and enzyme in a catalytic arrangement to drive self-anchored ubiquitination. Kiss, L., Clift, D., Renner, N., Neuhaus, D., James, LC. Nature Communications 12, 1220 https://doi.org/10.1038/s41467-021-21443-6
David’s group page
Leo’s group page
Previous Insights on Research
TRIM21 links antibody and T cell immunity to combat viral infection
Trim-Away: powerful new tool for studying protein function