 A research team from the LMB’s Cell Biology division, working with colleagues from the Structural Studies division, has revealed how cells are able to find and tag for degradation the partially synthesised proteins generated when ribosomes occasionally stall.
A research team from the LMB’s Cell Biology division, working with colleagues from the Structural Studies division, has revealed how cells are able to find and tag for degradation the partially synthesised proteins generated when ribosomes occasionally stall.
Cells make more than a hundred thousand new proteins every minute. Once in a while, one of the ribosomes making these proteins stalls, leaving an unfinished protein fragment. These incomplete proteins can interfere with normal cell functions, and over time, can cause neurodegeneration. To avoid this, cells have evolved a quality control system to tag these aberrant proteins for degradation. But how does this tagging machinery target only the rare faulty products without interfering with the vastly more abundant normal counterparts? Susan Shao and Manu Hegde have now provided an answer to this “Where’s Wally” puzzle.
In earlier work, the Hegde group had discovered how to carry out the tagging reaction in a test tube and found that Listerin is the enzyme that adds ubiquitins (the degradation tag) to the stalled protein. Exploiting this test tube system, Susan worked out the sequence of steps leading from a stalled ribosome to the Listerin-mediated ubiquitination of the unfinished protein. She discovered that two things happen before Listerin acts: the ribosome is split into its two subunits, and a new factor (called NEMF) binds the large subunit that contains the partially synthesised nascent protein. Only then is Listerin recruited, explaining why it does not ubiquitinate normal translating proteins.
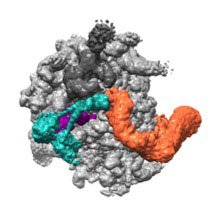
Susan then took advantage of recent cryo-EM developments, made in the LMB’s Structural Studies division, to solve the structure of a complex of NEMF and Listerin bound to a large ribosomal subunit housing an unfinished protein. The structure revealed that NEMF binds to the tRNA molecule attached to the end of the nascent protein. This tRNA is a telltale sign that protein synthesis has not finished and is normally buried inside an intact ribosome, which explains why the ribosomal subunits must split before NEMF binding. Listerin then binds NEMF and the ribosome, with its ligase domain right next to the nascent protein. Atomic models generated by Alan Brown, aided by sequence analysis by Balaji Santhanam, provided molecular insights into many of these interactions, which were then validated with mutational analyses.
Concepts from this new work may help understand other quality control pathways, where cells must ferret out occasional misfolded or damaged proteins amidst a sea of normal counterparts. Deficiencies in quality control underlie numerous human diseases and understanding this process may eventually lead to strategies aimed at avoiding accumulation of the aberrant proteins that accompany many forms of neurodegeneration.
This work was funded by the MRC and by St Johns College, Cambridge.
Further References:
Paper in Molecular Cell
Manu’s Group Page