 Recent work carried out by David Komander’s group in the LMB’s Protein and Nucleic Acid Chemistry Division, and published in two separate papers, has provided insight into human disease and the role played by ubiquitination, a process that affects many fundamental cellular processes.
Recent work carried out by David Komander’s group in the LMB’s Protein and Nucleic Acid Chemistry Division, and published in two separate papers, has provided insight into human disease and the role played by ubiquitination, a process that affects many fundamental cellular processes.
Work in David’s group aims to understand the cellular machinery for specific ubiquitin (Ub) assembly, disassembly, and binding on a structural and biochemical level. Ub forms at least eight types of polymers, and cells utilise this repertoire of modifications extensively in a range of signalling roles in diverse cellular processes, including protein degradation, the DNA damage response and the inflammatory response. Understanding these processes is key to understanding human diseases such as inflammatory diseases, cancer and early-onset Parkinson’s disease.
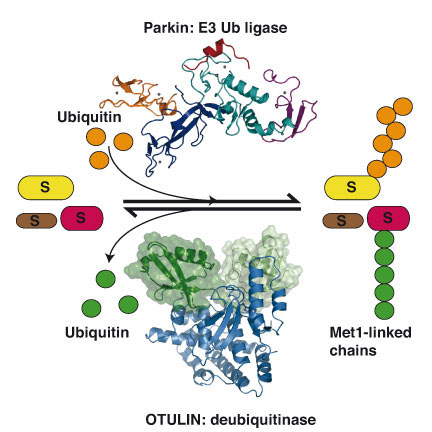
In the first paper, published in The EMBO Journal, work led by Tobias Wauer on the crystal structure of the protein Parkin is described. Parkin is a protein that belongs to the structurally uncharacterised RBR family of ubiquitin ligase enzymes. Mutations of this protein can lead to a hereditary form of early-onset Parkinson’s disease, an illness that affects many people worldwide and for which there is no cure. The structure explains how some of these mutations may affect the stability and function of the protein. The findings may in time reveal how the activity of Parkin is affected in patients and allow the development of compounds that alter Parkin activity, which could serve as ways to limit the progression and impact of Parkinson’s disease.
In the second paper, published in the journal Cell, work led by Kirstin Keusekotten and Paul Elliott, in collaboration with researchers from Cologne, Munich and Copenhagen, identified a new deubiquitinating enzyme in humans, OTULIN. This protein is shown to be the so far only enzyme to exclusively disassemble linear polyUb, an important Ub chain type that regulates inflammatory signalling pathways. To achieve its specificity, OTULIN is the first deubiquitinating enzyme to rely on substrate-assisted catalysis, utilising residues from the linear polyUb in the catalytic mechanism. The work further confirmed that linear Ub chains play key role in the nuclear factor κB (NF-κB) signalling pathway, which is often deregulated in inflammatory diseases and cancer. Identification of a new regulator for this pathway may enable new approaches to tackle many diseases.
This work was supported by the Medical Research Council, the European Research Council, the Lister Institute and the EMBO Young Investigator programme, the German Research Foundation, the Novo Nordisk Foundation and a Steno Fellowship from the Danish Council for Independent Research.
Further references
Article in The EMBO Journal on Parkin
Press release EMBO
Article in Cell on OTULIN
Preview article in Cell