Cells need to respond quickly when they encounter stress conditions in order to avoid consequences such as cell death. New research from Madan Babu’s group in the LMB’s Structural Studies Division has identified a mechanism by which cells can enhance the expression of stress-response genes by increasing the efficiency of protein synthesis specifically for these genes.
In order to make new proteins, genes are first transcribed to produce messenger RNA (mRNA) molecules, that are then translated into proteins. The information in the mRNA is encoded by 61 different nucleotide triplets known as codons. Codons are recognised by transfer RNAs (tRNAs) that carry amino acids, the individual components that make up proteins. As these 61 different codons encode for 20 different amino acids, most amino acids correspond to more than one codon. Some of these codons are more often used than others, for the same amino acid. It was previously noted that genes encoding growth proteins in the yeast Saccharomyces cerevisiae have more common codons, whereas genes for stress-response proteins tend to use more of the rare codons. This raises the question why cells use rare codons to make proteins that are needed under stress conditions.
Marc Torrent, the first author of this work from Madan’s group measured the abundance of all of the different tRNAs in yeast cells and how this changed in response to different stress conditions. They found that stress skews the total pool of different tRNAs so that there are more tRNAs for rare codons and fewer tRNAs for common codons. The team of scientists then demonstrated that changes in the abundance of tRNAs are sufficient to affect the rate of protein synthesis, as having more tRNAs available for rare codons will make translation of mRNAs containing a larger number of rare codons faster. As stress-response genes in yeast have a greater proportion of rare codons, the efficiency of translation of these mRNAs will be higher than for other mRNAs, and more stress-response proteins will be produced than other proteins when a cell is under stress conditions. Using these measurements, Guilhem Chalancon, joint first author of this work, designed proteins that contained identical amino acids, but used different, rarer, codons, resulting in better expression of the engineered proteins under stress conditions.
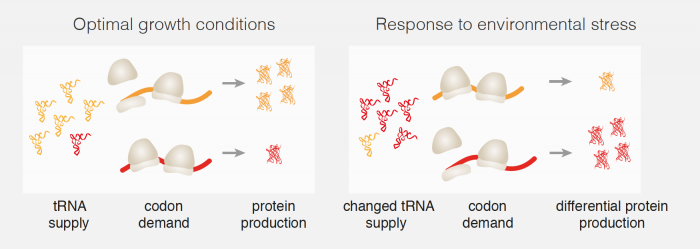
The process of decoding genes to produce proteins is highly conserved and similar in yeast and more complicated lifeforms, including humans. This means that this new role for tRNAs in the control of gene expression could have a role in human disease. For example, cancer is a disease in which genes that direct growth and cell division are expressed more than they should be, which would require abundance of the right tRNAs. Therefore, understanding how cells regulate the rates of synthesis of different proteins through control of the availability of tRNAs could point to new directions for cancer treatment.
This work was funded by the MRC, the Lister Institute Research Prize, Marie Curie Actions, the Ministerio de Economía y Competitividad, and the Gates Cambridge Scholarship.
Further references:
Paper in Science Signaling
Science Signaling Focus article: “Coping with stress by regulating tRNAs”
Madan Babu’s group page