Leveraging next generation sequencing technology, deep screening promises to accelerate antibody discovery for a wide range of targets by generating sequence-function correlation data on an unprecedented scale
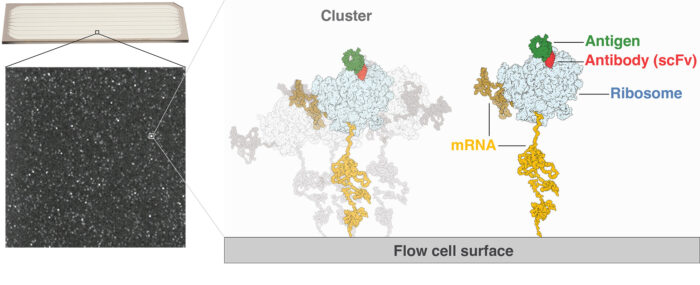
Therapeutic antibodies have had a transformative clinical impact, notably in inflammatory diseases and cancer, but their development remains challenging, time-consuming, and costly. Now Philipp Holliger and Ben Porebski in the LMB’s PNAC Division, in collaboration with a team from the AstraZeneca Biologics Engineering group, have developed a new, ultra-high-throughput method that enables rapid discovery of clinically relevant antibodies.
The new method builds upon the Illumina HiSeq next generation sequencing platform, combining massively parallel clustering and sequencing of antibody genes, ribosome display and rapid affinity screening in the order of 100 million individual antibody-antigen interactions all within a single instrument. In just three days, antibody leads can be identified from a library, resulting in one case in an over 2000-fold improvement in affinity and over 35,000-fold increase in potency. This is a dramatic acceleration upon previous methods which took months to reach the same goal.
The output of a single deep screening experiment is a large antibody-antigen binding correlation dataset, which includes information on which sequences bind or do not bind to antigens, and with what affinity. The team also showed that these vast datasets can be used to train a machine learning model, specifically a large language model, to “speak antibody”, that is to implicitly “understand” what makes an antibody functional. Once trained, the language model is then able to generate novel and higher-affinity antibody sequences that encode functional and potentially superior antibodies not found in nature, thus further accelerating antibody discovery.
This new high-throughput method has the potential to transform the discovery of antibodies for medicine from an arduous process of multiple rounds of looking for a needle in a haystack to a simple survey of all antibodies in a given library (the whole haystack) in a single experiment. By reducing both time and cost of new antibody drug discovery for a wide range of disease targets, deep screening has the potential to have significant impact.
The development of deep screening builds upon the LMB’s previous pivotal antibody technology research: the discovery of a method to produce monoclonal antibodies by César Milstein and Georges Köhler, and production of humanised and entirely human antibodies by Greg Winter and Michael Neurberger.
This work was funded by the LMB’s and AstraZeneca’s Blue Sky Collaboration (a research collaboration between AstraZeneca UK Limited and the Medical Research Council, reference BSF24) and supported by UKRI MRC and EMBO.
Further references
Rapid discovery of high-affinity antibodies via massively parallel sequencing, ribosome display, and affinity screening. Porebski, BT., Balmforth, M., Browne, G., Riley, A., Jamali, K., Fürst, MJLJ., Velic, M., Buchanan, A., Minter, R., Vaughan, T., Holliger, P. Nature Biomedical Engineering
Philipp’s group page
Previous Insight on Research articles
Engineered polymerase enzyme presents new opportunity for quick and efficient modified nucleic acid synthesis
A new directed evolution technique to unlock the potential of XNAs