Single-cell RNA sequencing of suprachiasmatic nucleus cells identifies key cell populations and reveals the role of the neuropeptide Prokineticin to circadian governance
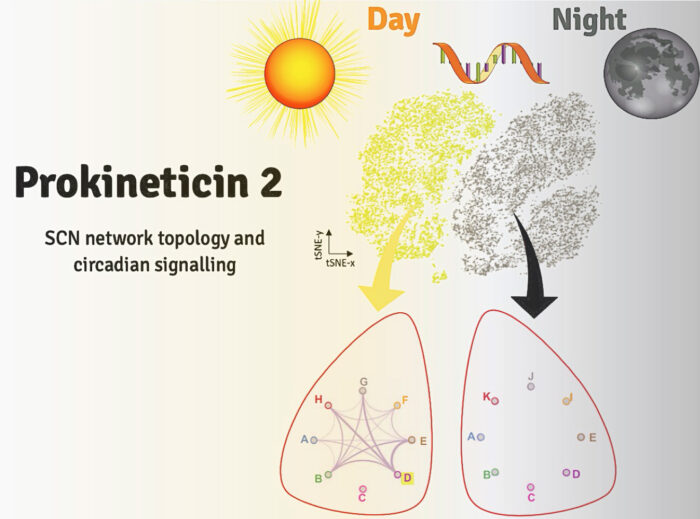
Circadian rhythms dominate our daily lives influencing for example, when we feel tired or hungry, when we are most likely to be able to focus attention and concentrate and when we are likely to perform best in physical exercise. In mammals, circadian rhythms are governed by the suprachiasmatic nucleus (SCN), located in the hypothalamus of the brain. The 20,000 clock cells of this principal pacemaker synchronise cellular clocks in individual cells across the body to maintain daily rhythms of physiology. However, the topology and the specific roles of the distinct cell populations within the SCN which direct its network functions are not well understood. Now, Michael Hastings’ group, in the LMB’s Neurobiology Division, have sequenced, at a single cell level, the transcriptomes – messenger RNA molecules expressed by active genes – within the SCN in day and in night in order to neurochemically characterise the features necessary for circadian function. The study has led the group to identify the neuropeptide Prokineticin 2 (Prok2) and its signalling target receptor Prokineticin Receptor 2 (ProkR2) as key drivers of the circadian network.
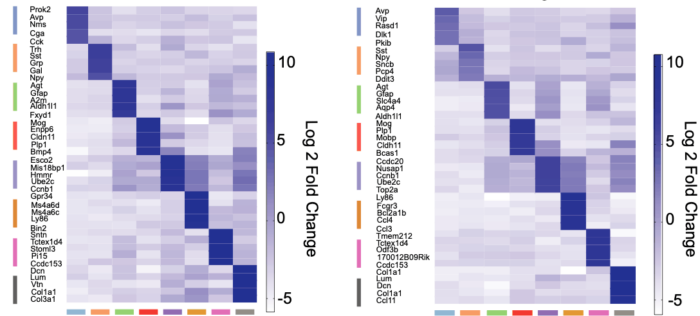
To identify the SCN’s composition of cells and network structure, group member and lead author Emma Morris conducted single-cell RNA sequencing (scRNAseq) of organotypic SCN slices from mice, which contained the full apparatus for autonomous molecular time-keeping. Taking samples at different times across the circadian clock allowed the group to examine the patterns of gene activation in different cell types within the SCN. In total, the group harvested over 30,000 cells from multiple slices, which were processed using scRNAseq and classified into eleven distinct neuronal sub-populations.
These tests exposed how the signalling network across the SCN changes over day and night. Specifically, genes associated with cell contacts and synaptic function were significantly up-regulated during the circadian day, whilst circadian night showed increased protein degradation and a loss of neuropeptide transport. The most dramatically up-regulated gene was Prok2 which increased fivefold during the day. Further, the signalling between Prok2 and its target ProkR2 displayed circadian characteristics distinct from other signalling molecules. Using advanced microscopy, the group then showed that the Prok2 and ProkR2 sub-populations of cells (which comprise only about 20% of the SCN in total), are able to determine the ensemble period for SCN and to initiate circadian oscillations, making them central controllers of the entire network.
This research has advanced understanding of the SCN by identifying the role of specific cellular populations, further defining its network topology and illustrating how this is remodelled across circadian time. Identifying the key roles played by Prok2 and ProkR2 improves our understanding of what makes the SCN such a powerful time-keeper and how our body clocks are controlled. The regulation of almost all of our physiological processes can be traced back to the SCN. In addition to regulating behaviours such as appetite and ability to concentrate, circadian rhythms are also implicated in health and disease, for instance the worsening of asthma symptoms during the night or the increased risk of heart attacks early in the morning. The increased understanding of the functional components of the SCN will hopefully contribute to the development of diagnostic markers and therapeutic interventions by exposing targetable signalling pathways.
This work was funded by UKRI MRC and BBSRC.
Further references
Single-cell transcriptomics of suprachiasmatic nuclei reveal a Prokineticin-driven circadian network. Morris, EL., Patton, AP., Chesham, JE., Crisp, A., Adamson, A., Hastings, M.H., EMBO Journal 40:e108614