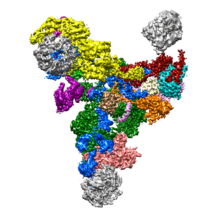
 Researchers in the LMB’s Structural Studies Division have been able to show in more detail than ever before the structure of a large part of the spliceosome, a macromolecular machine involved in the maturation of messenger RNAs for protein synthesis. Thi Hoang Duong (Kelly) Nguyen, Wojciech Galej, Xiao-Chen Bai, Andy Newman and Chris Oubridge in Kiyoshi Nagai’s and Sjors Scheres’ laboratories, used electron cryo-microscopy (cryo-EM) to map out an accurate three-dimensional picture of the RNA-protein complex, called the U4/U6.U5 tri-snRNP.
Researchers in the LMB’s Structural Studies Division have been able to show in more detail than ever before the structure of a large part of the spliceosome, a macromolecular machine involved in the maturation of messenger RNAs for protein synthesis. Thi Hoang Duong (Kelly) Nguyen, Wojciech Galej, Xiao-Chen Bai, Andy Newman and Chris Oubridge in Kiyoshi Nagai’s and Sjors Scheres’ laboratories, used electron cryo-microscopy (cryo-EM) to map out an accurate three-dimensional picture of the RNA-protein complex, called the U4/U6.U5 tri-snRNP.
In eukaryotes protein coding genes are segmented. Genes are copied into messenger RNA precursors (pre-mRNAs) and in order for messenger RNAs to be translated into proteins, the pre-mRNAs must be cut into pieces and small non-coding segments, called introns, removed. The spliceosome recognises and removes introns from the pre-mRNA and stitches coding segments (exons) together. Due to the complex nature of this molecular machine comprising 30 proteins and 3 RNAs and the structural flexibilities previous attempts to visualise it by cryo-EM have been limited to low resolution. A breakthrough came last year when an outline structure of the U4/U6.U5 tri-snRNP was revealed by the same scientists through combining cryo-EM pictures with known crystal structures of its building blocks. Emboldened by these results, the scientists purified minute amounts of the 1.5 MDa U4/U6.U5 tri-snRNP complex from hundreds of litres of yeast culture. They then took thousands of cryo-EM images to increase the number of single particles of this complex they could analyse. By using image-processing software developed by Sjors to remove the fuzzy ‘noise’ from the images, they succeeded in visualising a clear picture the U4/U6.U5 tri-snRNP, including the flexible sections important in activation of the Spliceosome, to the atomic level.
By further analysing versions of U4/U6.U5 tri-snRNP mutated by the group to change certain amino acids into others, a model mechanism for the activation of the spliceosome was proposed.
This work has provided a crucial step towards understanding the shape of the spliceosome and how it functions. Malfunction of this machine is implicated in diseases including degenerative eye disease, retinitis pigmentosa, and some cancers. In future, it may be possible to understand how specific mutations in the spliceosome lead to these diseases.
The work was funded by the MRC.
Further references:
Paper in Nature
Previous Insight on Research article
Kiyoshi’s Group Page
Sjors’ Group Page