Synthetic biological methodology reveals regulatory role of cryptochrome clock proteins in maintaining daily rhythms within the suprachiasmatic nucleus
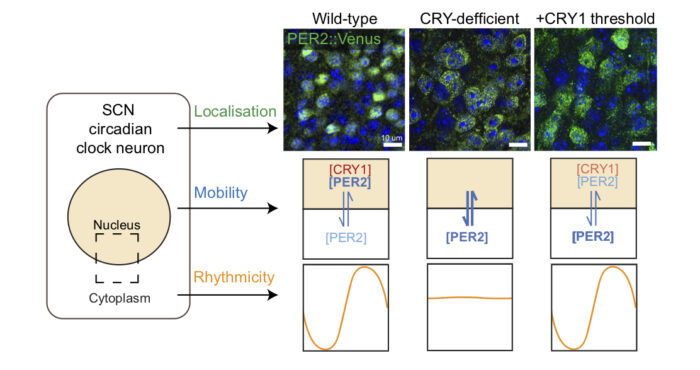
In mammals, daily physiological and behavioural rhythms are governed by an internal, molecular clockwork, which is present in almost all cells of the body. This circadian rhythm is coordinated by the suprachiasmatic nucleus (SCN) – the ‘master clock’ in the brain – which is comprised of roughly 20,000 cells. SCN timekeeping relies upon transcriptional/translational feedback loops (TTFLs) in which PERIOD (PER) and CRYPTOCHROME (CRY) clock proteins negatively regulate the circadian rhythm by associating and translocating in the nucleus to inhibit their own expression. Currently, in depth understanding of the body’s circadian clock is curtailed by a lack of knowledge surrounding the individual and interactive behaviours of PER and CRY proteins, and their guiding mechanisms. A new study from Michael Hastings’ group in the LMB’s Neurobiology Division, in collaboration with Andrew Loudon at the University of Manchester, utilised advanced synthetic biological methodology, pioneered by Jason Chin in the LMB’s PNAC Division, to identify CRY proteins as the key factor to maintaining circadian functionality.
Nicola Smyllie, the study’s lead author, used microscopy to examine the behaviour of the protein PER2 within living cells of the SCN. Real-time imaging of endogenous PER2 revealed that the protein is predominantly nuclear and operates at three levels of mobility; fast, slow, and immobile (the latter being the most prevalent). Additionally, Nicola noted that when comparing mobile PER2 proteins to other transcriptional regulators, PER2 is notably more itinerant, both within and between cellular compartments such as the nucleus and cytoplasm.
To determine the role of CRY, Nicola used genetically modified brain tissue that lacked CRY proteins and used viral vectors to ‘add back’ different forms of CRY to the tissue and measure the impact on PER2 protein behaviour.
In CRY-deficient brain tissue, the group observed that rather than being predominantly nuclear, expression of PER2 was significantly more cytoplasmic, and thus impaired the rhythmic clock function of the SCN. Furthermore, in this modified tissue, PER2 was found to be more mobile; the individual PER2 molecules categorised as ‘fast’ were even faster and there was a markedly lower proportion of immobile molecules.
The group used translational switching to “dial in” the amounts of added CRY1 incrementally. Interestingly, the study found that the presence of very low levels of CRY1 was sufficient to achieve a critical threshold of CRY and partially relocate PER2 to the nucleus. This allowed the resumption of accurate timekeeping – as observed in the wild type tissue. These results highlight the unexpected efficacy of relatively low levels of nuclear CRY1 for the regulation of PER nuclear localisation, and thereby the maintenance of TTFLs in the SCN.
These findings offer the first insight into the relationship between molecular and cellular processes that govern the SCN, and therefore control bodily circadian rhythms in mammals. This holds important clinical implications, as the disruption of circadian rhythms, such as that experienced amongst shift workers, has long been linked to a range of metabolic, cardiovascular and neurological diseases and disorders. By continuing to develop our understanding of circadian rhythms, we can better understand the causes of such complex, systemic diseases and implement novel therapeutic responses, such as using chronotherapy to optimise delivery time of drug treatments to maximise efficacy.
This work was funded by UKRI MRC and UKRI BBSRC.
Further references
Cryptochrome proteins regulate the circadian intracellular behavior and localization of PER2 in mouse suprachiasmatic nucleus neurons. Smyllie, NJ., Bagnall, J., Koch, AA., Niranjan, D., Polidarova, L., Chesham, JE., Chin, JW., Partch, CL., Loudon, ASI., Hastings, MH. PNAS Vol. 119 No. 4
Michael’s group page
Jason’s group page
Andrew Loudon, University of Manchester
Previous Insight on Research articles
Advanced understanding of how suprachiasmatic nucleus regulates the body’s circadian rhythm
A cellular pacemaker in the central body clock
A new tool using genetic code expansion to study circadian rhythms