New improved polymerase unlocks possibility to solve slow and costly approach of synthesising modified nucleic acids, opening up potential for exploration of vast numbers of candidate sequences
Nucleic acids are hugely important clinically, as their unique capability of finding and pairing with a specific, complementary sequence means they are easily targeted therapeutically once a pathological sequence is known. However, in addition to pathogenic nucleic acid material, natural host DNA and RNA are susceptible to degradation. This occurs naturally through nucleases—a type of host enzyme which breaks down nucleic acids, including severing intruding nucleic acids from pathogens.
Certain nucleic acids not found in nature have been shown to possess pharmacologically interesting properties, such as high biostability and resistance to our body’s degradative defence mechanisms. These xeno-nucleic acids (XNAs) are genetic polymers, like DNA or RNA, but contain altered sugar rings, bases, or backbones.
In recent years, XNAs have been used to develop novel drugs, including the first approved medicine for spinal muscular atrophy. Thus far, the process to synthesise candidate drugs has been slow and costly, making large-scale explorations of libraries of candidate molecules near impossible. Now, Philipp Holliger’s group in the LMB’s PNAC Division have developed a polymerase enzyme which could ease this problem.
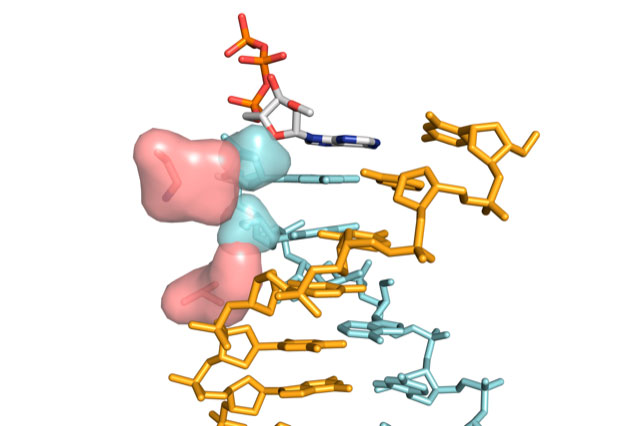
Lead authors Niklas Freund (a PhD student in Philipp’s group) and Alexander Taylor (formerly a Postdoc in Philipp’s group, now a Group Leader at the Cambridge Institute of Therapeutic Immunology and Infectious Disease) designed and modified this enzyme after closely studying a related three-dimensional protein structure. They observed clear clashes between the 2’-modified nucleic acid structure and the interior of the protein. In the polymerase enzyme, they identified two ‘gatekeeper’ positions. The group mutated these regions to allow for free passage of large, modified nucleic acids. Capability tests of this improved enzyme found that it is substantially faster than previous engineered ‘progenitor’ or ‘parent’ polymerases, while retaining equal fidelity for making XNAs.
The group then used this engineered polymerase enzyme to produce modified nucleic acid structures capable of selectively cutting messenger RNAs (mRNAs) that contain specific, single changes causing diseases, such as many cancers. They also created structures that fold into three-dimensional shapes which, like antibodies, are capable of binding a protein surface with high affinity.
This newly engineered polymerase enzyme should allow for greater exploration of the benefits of using unnatural nucleic acids, combined with their conserved mode of action. This technology would advance the development of next-generation therapeutic agents, which could fulfil the promise of a currently under-developed therapeutic mode of action.
This work was funded by UKRI MRC, Boehringer Ingelheim Fonds, Flanders Research Foundation Fund of Scientific Research, KU Leuven, National Institutes of Health and supported through the LMB’s and AstraZeneca’s BlueSky collaboration (a research collaboration between AstraZeneca UK Limited and the Medical Research Council, reference BSF1 and BSF11).
Further references
A two-residue nascent strand steric gate controls synthesis of 2’-O-methyl- and 2’-O-(2-methoxyethyl)-RNA. Freund, N., Taylor, AI., Arangundy-Franklin, S., Subramanian, N., Peak-Chew, S-Y., Whitaker, AM., Freudenthal, BD., Abramov, M., Herdewijn, P., Holliger, P. Nature Chemistry
Philipp’s group page
Alexander Taylor’s group page
Blue Sky Collaboration
Previous Insight on Research articles
A new directed evolution technique to unlock the potential of XNAs
Uncharged DNA-like molecules can store genetic information and function like antibodies