Bacterial proteins Vipp1 and PspA are found to be members of a family of membrane-remodelling proteins once thought to be exclusive to eukaryotic cells
Structure of a cyanobacterium Vipp1 polymer. Each colour corresponds to one rung or layer containing 14 Vipp1 subunits. The polymer has 84 subunits and a molecular weight of ~2.4 megadaltons. © Harry Low
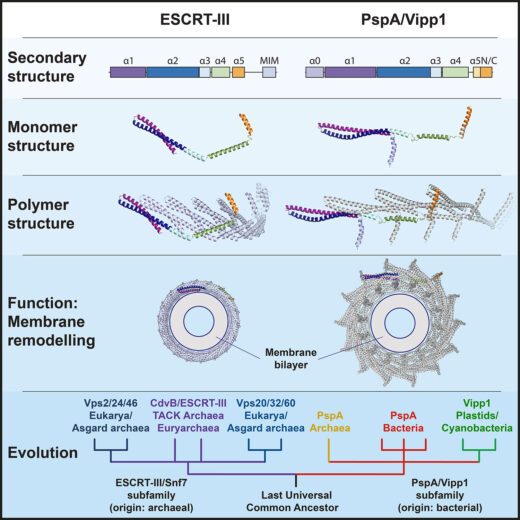
Membrane remodelling and repair are essential for all cells, with cell division, and in particular the cutting of the membrane to separate the two daughter cells, being a universal concern for all organisms. One major class of proteins that control this are ESCRT-III proteins, which had once been thought to be exclusively found in eukaryotes; organisms whose cells contain a nucleus, ranging from unicellular yeast to complex plants and animals. However, Buzz Baum’s group, in the Cell Biology Division, working with Harry Low’s group at Imperial College London, have shown that this family of proteins has relatives in all domains of life, including bacteria.
Life can be divided into three branches, eukaryotes, archaea, and bacteria. Archaea and bacteria are both prokaryotes, meaning that they lack a nuclear membrane-enclosed nucleus, but archaea are more closely related to eukaryotes than they are to bacteria. Although they were once thought to be defining features of eukaryotic cells, there are a number of cytoskeletal proteins and membrane remodelling systems that are now known to have relatives in archaea and bacteria. Similarly, ESCRT-III homologues had been discovered in archaea, leading to the suggestion that they might have originated in the ancestors of archaea and eukaryotes but, working with their collaborators, Buzz’s group has now shown these cytoskeletal elements are much older in origin.
ESCRT-III (Endosomal Sorting Complexes Required for Transport) proteins form polymers that are involved in a number of cell biological processes, including formation of vesicles for processing of ubiquitin-tagged proteins, membrane repair, nuclear envelope reformation, and viral budding. To perform this wide range of membrane remodelling functions, ESCRT-III polymers can form a variety of different structures, such as helical tubes, rings, spirals, filaments, and cones. Understanding of their function will benefit from insights into how these different structures can be self-organised and allow different activities.
Diorge Souza, a postdoctoral scientist in Buzz’s group, first observed the similarity of a particular bacterial protein family, PspA/Vipp1, when searching for ESCRT-III homologues. Computational predictions based on this sequence similarity then showed him that the similarity likely extended to the 3D structure and potential formation of polymers. To investigate this further, Buzz and Diorge collaborated with Harry Low, a former PhD student and postdoc in the LMB’s Structural Studies Division and now Senior Research Fellow at Imperial. Jiwei Liu, a member of Harry’s group, was able to solve the Vipp1 structure using cryo-EM.
Together, their work showed that flexibility in the Vipp1 monomer enables it to polymerise to form rings over a range of symmetries. Each of these rings are built from rungs that stack and spontaneously self-organise to form domes, the inside of which is able to bind and deform membranes. This demonstrates comparable points of flexibility to eukaryotic ESCRT-III proteins and suggested that there are conserved mechanistic principles underlying the functions of all of these related proteins across all domains of life.
This work shows that the last common ancestor of everything alive today was a relatively complex organism, possessing ancestral forms of many proteins that had once been thought to be later developments, thereby demonstrating the universality of how life works and how basic cellular processes are controlled.
The work was funded by UKRI MRC, BBSRC, the Wellcome Trust and the Royal Society.
Further references
Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. Liu, J., Tassinari, M., Souza, DP., Naskar, S., Noel, JK., Bohuszewicz, O., Buck, M., Williams, TA., Baum, B., Low, HH. Cell 184: 1-14
Buzz’s group page
Harry Low’s page