Buzz Baum
The generation and evolution of biological form: from archaea to eukaryotes
bbaum@mrc-lmb.cam.ac.ukPersonal group site
My team is interested in the generation of biological form or “morphogenesis”. We study events at the cellular scale because the cell is the unit of life. Much of the lab’s work focuses on cell division – the process by which becomes two. This is one of the most remarkable and fundamental features of all life on earth. During division, within a timeframe of a few minutes, all the component parts of a cell must be moved apart and partitioned into two daughter cells. Thus, division must be done with precision to ensure the propagation of a lineage, and errors in division are frequently associated with diseases like cancer in humans and can cause cell death.
Rapid changes in cell shape and organisation, including those that accompany division, are brought about by a network of dynamic polymers – the cytoskeleton. These filaments convert chemical energy into force as they grow and shrink. Strikingly, recent evolutionary studies have shown that the key cytoskeletal elements used to drive division and other shape changes in our cells are ancient proteins, which we inherited from our archaeal ancestors during the process of eukaryogenesis.
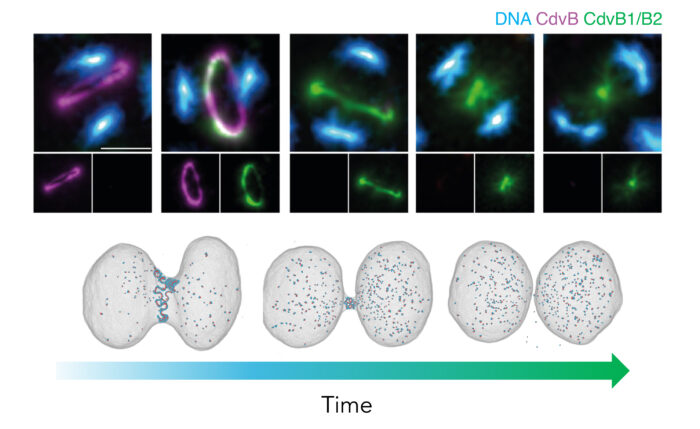
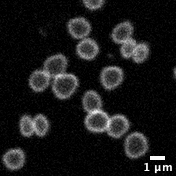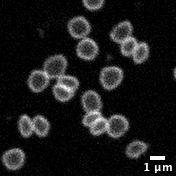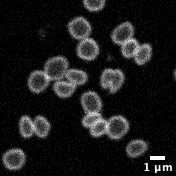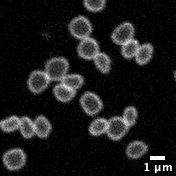
Taking advantage of this discovery, in exploring the fundamentals of cell division, my lab is looking at the similarities and differences in the way archaea and eukaryotic cells divide. Using Sulfolobus as a genetically tractable model in which to study archaeal cell biology, we aim to identify conserved processes, core principles, and to determine how division has changed during the transition from archaea to eukaryotes. This work has led us to study the structure and function of ESCRT-III filaments, which catalyse the final step in division in both Sulfolobus and in human cells.
Working with archaea also means grappling with some the weirder features of these extremophiles. Sulfolobus cells, which were originally isolated from hot acidic pools in Yellowstone National Park, grow at 75°C and have membranes that are largely composed of monolayer lipids. Thus, understanding their cell biology requires re-thinking some of our most basic assumptions (bilayers membranes) and the development of new research tools and methods (high-temperature live cell imaging).
Taking this evolutionary cell biology approach further, in the coming years we will also be turning our attention to the Asgard archaea. These are our closest living archaeal relatives. Asgard archaea have extraordinary shapes and, in some cases, partner with cells of other species to grow. They are therefore ideal models in which to study the evolution of cell division, cell shape control, and symbiosis.
We expect this work, much of which is done with collaborators at the LMB, in the UK and overseas, to shed light on the process by which eukaryotic cells emerged from the symbiosis of an Asgard archaeal cell with an alpha-proteobacterial partner that gave rise to the mitochondria. In doing so, we will put the “inside-out model” for the origin of the eukaryote cell to the test. For the moment, the evolution of eukaryotes remains one of the greatest mysteries in the history of life on earth. We hope to help change this.
Selected Papers
- Cezanne A, Hoogenberg B, Baum B. (2023)
Probing archaeal cell biology: exploring the use of dyes in the imaging of Sulfolobus cells
Frontiers in Microbiology 14: - van Wolferen M, Pulschen AA, Baum B*, Gribaldo S* Albers SV (2023)
The cell biology of archaea
Nature microbiology 7 (11): 1744-1755 - Hurtig F, Burgers TCQ, Cezanne A, Jiang X, Mol FN, Traparić J, Pulschen AA, Nierhaus T, Tarrason-Risa G, Harker-Kirschneck L, Löwe J, Šarić A, Vlijm R, Baum B (2023)
The patterned assembly and stepwise Vps4-mediated disassembly of composite ESCRT-III polymers drives archaeal cell division
Sci Adv. 17;9(11): eade5224 - Hatano T, Palani S^, Papatziamou D^, Salzer R^, Souza DP^, Tamarit D^, Makwana M, Potter A, Haig A, Xu W, Townsend D, Rochester D, Bellini D, Hussain HMA, Ettema TJG, Löwe J, Baum B*, Robinson NP* & Balasubramanian M* (2022)
Asgard archaea shed light on the evolutionary origins of the eukaryotic ubiquitin-ESCRT machinery
Nat Communications 13(1): 3398. - J Liu, M Tassinari, D P Souza, S Naskar, JK Noel, O Bohuszewicz, M Buck, TA Williams, B Baum*, HH Low* (2021)
Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodelling superfamily.
Cell 184 (14): 3660-3673. - Roubinet C, White IJ, & Baum B (2021)
Asymmetric nuclear division in neural stem cells generates sibling nuclei that differ in size, envelope composition, and chromatin organization
Current Biology 31(18): 3973-3983 - AK Pfitzner, V Mercier, X Jiang, J Moser von Filseck, B Baum, A Šarić, A. Roux (2020)
Sequential polymerization of ESCRT subunits drives membrane deformation and fission
Cell 182: 1140-1155. - G Dey, S Culley, S Curran, U Schmidt, R Henriques, W Kukulski, and B Baum. (2020)
Closed mitosis requires local disassembly of the nuclear envelope.
Nature 585: 119-123. - G Tarrason Risa, F Hurtig, S Bray, AE Hafner, L Harker-KirschnecK, P Faull, C Davis, D Papatziamou, DR Mutavchiev, C Fan, L Meneguello, AA Pulschen, G Dey, S Culley, M Kilkenny, L Pellegrini, RAM de Bruin, R Henriques, AP Snijders, A Šarić, A-C Lindås, NP Robinson* and B Baum* (2020)
The proteasome controls ESCRT-III-mediated cell division in an archaeon.
Science 369: (6504). - AA Pulschen, DR Mutavchiev, S Culley, KN Sebastian, J Roubinet, M Roubinet, G Tarrason Risa, M van Wolferen, C Roubinet, U Schmidt, G Dey, S-V Albers, R Henriques, B Baum. (2020)
Live Imaging of a Hyperthermophilic Archaeon Reveals Distinct Roles for Two ESCRT-III Homologs in Ensuring a Robust and Symmetric Division.
Current Biology 30: 1-8. - HK Matthews, S Ganguli, K Plak, AV Taubenberger, M Piel, J Guck, B Baum. (2020)
Oncogenic signaling alters cell shape and mechanics to facilitate cell division under confinement.
Dev Cell 52(5): 563-573. - Rodrigues, NT., Lekomtsev, S., Jananji, S., Kriston-Vizi, J., Hickson, GR., & Baum, B. (2015)
Kinetochore- localized PP1-Sds22 couples chromosome segregation to polar relaxation.
Nature 524(7566): 489-492. - Rohn JL., Patel JC., Neumann B., Bulkescher J., Mchedlishvili N., McMullan RC., Quintero OA., Ellenberg J, & Baum B (2014)
An Actin-Based Myosin Motor, Myo19, Couples Mitochondrial Segregation to Cell Division
Current Biology 24: 2598- 2605 - Baum, D, Baum, B. (2014)
An inside-out origin for eukaryote cells.
BMC Biol. 12: 76.
Group Members
- Alice Cezanne
- Sherman Foo
- Yin-Wei (Kris) Kuo
- Magda Lechowska
- Fraser MacLeod
- Florian Mayer
- Joe Parham
- Arthur Radoux
- Iain Richard
- Jeanne Salje