

Precise chromosome replication is essential for maintaining genome stability, yet the process itself is inherently risky. Duplex DNA must be transiently separated into unstable single-strands; a vast number of DNA bases must be accurately copied; and replication forks frequently encounter protein barriers and unrepaired DNA damage that can stall and even derail the replication machinery (replisome).
Our lab is interested in mechanisms of chromosome replication, and how these mechanisms contribute to faithful genome duplication.
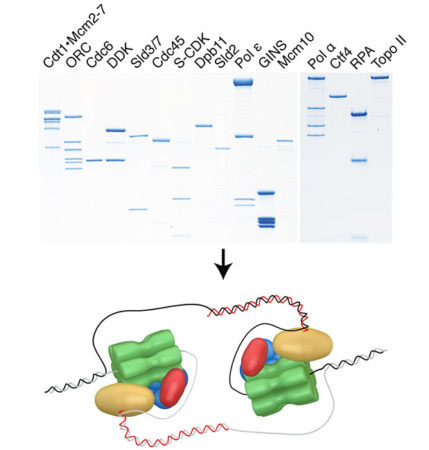
The eukaryotic replisome is a highly complex molecular machine that is assembled from in excess of 30 gene products and which orchestrates the activities of multiple enzymatic components. To study this remarkable machine in action our lab reconstitutes replication in vitro using purified proteins. This highly defined experimental system, used in conjunction with genetic approaches and replication proficient cell-free extracts, enables us to investigate fundamental aspects of replisome biology.
Selected Papers
- Yasemin Baris, Martin R. G. Taylor, Valentina Aria & Joseph T. P. Yeeles (2022)
Fast and efficient DNA replication with purified human proteins
Nature, 10.1038/s41586-022-04759-1. - Jenkyn-Bedford, M., Jones, M.L., Baris, Y., Labib, K.P.M., Cannone, G., Yeeles, J.T.P., and Deegan, T.D. (2021)
A Conserved Mechanism for Regulating Replisome Disassembly in Eukaryotes.
Nature, 10.1038/s41586-021-04145-3. - Jones, M.L., Baris, Y., Taylor, M.R.G., and Yeeles, J.T.P. (2021)
Structure of a human replisome shows the organisation and interactions of a DNA replication machine.
EMBO J., e108819. https://doi.org/10.15252/embj.2021108819 - Baretić, D., Jenkyn-Bedford, M., Aria, V., Cannone, G., Skehel, M., Yeeles, JTP. (2020)
Cryo-EM structure of the Fork Protection Complex bound to CMG at a replication fork.
Molecular Cell 78: 1-15. - Guilliam, TA., Yeeles, JTP. (2020)
Reconstitution of translesion synthesis reveals a mechanism of eukaryotic DNA replication restart.
Nature Structural and Molecular Biology, Epub ahead of print. - Aria V, Yeeles JTP. (2018)
Mechanism of Bidirectional Leading-Strand Synthesis Establishment at Eukaryotic DNA Replication Origins.
Mol Cell. 73: 199-211 e10. - Taylor MRG, Yeeles JTP. (2018)
The Initial Response of a Eukaryotic Replisome to DNA Damage.
Mol Cell. 70(6): 1067-1080. - Yeeles JTP, Janska A, Early A, Diffley JFX. (2017)
How the Eukaryotic Replisome Achieves Rapid and Efficient DNA Replication.
Mol Cell. 65(1): 105-116. - Yeeles, J.T., Deegan, T.D., Janska, A., Early, A. and Diffley, J.F. (2015)
Regulated eukaryotic DNA replication origin firing with purified proteins.
Nature 519(7544): 431-435.
Group Members
- Federico Fassetta
- Emma Fletcher
- Morgan Jones
- Harry Orrin
- Cristian Polo Rivera
- Jonas Roske
- Eleanor Taylor-Cross