

Harvey McMahon
Membrane curvature as an organizing principle for eukaryotic cell biology
Personal group site
Cell shape is adapted to function. Organelle shape and local membrane architectures are likewise optimised for the processes that take place on and within these microenvironments. We focus on the dynamic regulation of membrane shape, which can occur by the interplay between the transient and regulated insertion of membrane bending motifs and the detection and stabilisation of membrane shape.
This approach has allowed us not only to describe the biophysics of membrane shape changes but also to take a fresh look at physiological processes like exocytosis and endocytosis. In doing so we have noted that proteins with amphipathic helices or hydrophobic membrane-inserting loops are likely to effect or respond to curvature and that the membrane interaction surfaces of proteins can sense shape (like proteins of the BAR superfamily).
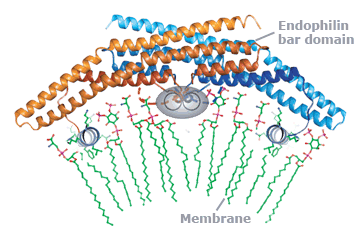
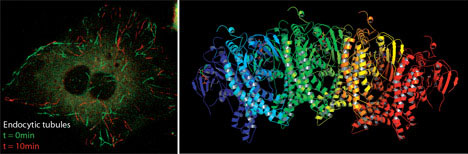
superfamily member which remodels membranes.
This molecular view has allowed us to ascribe novel cell-biological functions to proteins (e.g. the mechanistic affect of synaptotagmin in membrane fusion, and the role of endophilin in a non-clathrin pathway of endocytosis) and to give a more insightful view of how these processes work. Thus we can now go from the biophysics of a molecule, to better understanding of known pathways and to the molecular characterisation of novel cellular trafficking pathways both of endocytosis and exocytosis.
Selected Papers
- Almeida-Souza, L., Frank, R.A.W., García-Nafría, G., Colussi, A., Gunawardana, N., Johnson, C.M., Yu, M., Howard, G., Andrews, B., Vallis, Y., McMahon, H.T. (2018)
A Flat BAR Protein Promotes Actin Polymerization at the Base of Clathrin-Coated Pits
Cell 174(2): 325-337. - Boucrot, E., Ferreira, A.P.A., Almeida-Souza, L., Debard, S., Vallis, Y., Howard, G., Bertot, L., Sauvonnet N. and McMahon, H.T. (2015)
Endophilin marks and controls a clathrin-independent endocytic pathway.
Nature 517: 460-465. - Martens, S., Kozlov, M.M. and McMahon, H.T. (2007)
How synaptotagmin promotes membrane fusion.
Science 316: 1205-1208. - McMahon, H.T. and Gallop, J.L. (2005)
Membrane curvature and mechanisms of dynamic cell membrane remodelling.
Nature 438: 590-596. - Boucrot, E., Pick, A., Çamdere, G., Liska, N., Evergren, E., (McMahon, H.T. and Kozlov, M.M.) (2012)
Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains.
Cell 149: 124-136. - Henne, W.M., Boucrot, E., Meinecke, M., Evergren, E., Vallis, Y., Mittal R. and McMahon, H.T. (2010)
FCHo Proteins are Nucleators of Clathrin-Mediated Endocytosis.
Science 328: 1281-1284. - Daumke, O., Lundmark, R., Vallis, Y., Martens, S., Butler, P.J.G. and McMahon, H.T. (2007)
Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling.
Nature 449: 923-927.
Group Members
- Mahmoud Bassal
- Ting-Mei Liao
- Rohit Mittal
- David Paul