

The ability to watch development as it unfolds reveals how tissues form, however mammalian development has been notoriously difficult to visualize. Embryos are highly sensitive to environmental and culture conditions when removed from the mother, and in addition are extremely photosensitive.
We have developed an advanced light-sheet microscope to gently and comprehensively image mouse embryo development at single-cell resolution over a course of days. With this system and a suite of computational tools, we follow changes in cell fate, visualize the organization of tissue structures, and measure the forces involved in shaping those structures. Starting with the endoderm, we are examining how and where forces are generated to turn a relatively simple, single cell layered structure into the primordial gut tube, as well as how this process is coordinated with the development of neighbouring germ layers such as the heart tube and neural ectoderm.
One of the ultimate aims of the lab is to generate a comprehensive force-map of development in the early embryo that, when coupled with our existing knowledge of gene expression and cell fate maps, will allow us to generate computational models that can be used to design and engineer better 3D culture or synthetic tissue culture systems, enabling us to grow more complex structures in a dish and one day build working tissues and organs.
Two-day long time-lapse imaging of an H2BeGFP expressing mouse embryo from E6.5 d.p.c. to E8.5 d.p.c. (Top panel: distal view, bottom left panel: AP view, bottom right panel: lateral view).

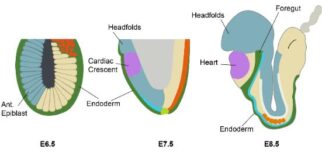
Time-lapse imaging of early cardiac development using an early
cardiac marker (Nkx2.5, in green) and a ubiquitous label (membrane
marker, in magenta). The cardiac crescent begins to come together
over the invaginating foregut pocket before converging at the midline
and forming the early heart tube.
Selected Papers
- McDole K., Guignard L., Amat F., Berger A., Malandain G., Royer LA., Turaga SC., Branson K. & Keller PJ. (2018)
In toto imaging and reconstruction of post-implantation mouse development at the single-cell level.
Cell 175: 859-876. - McDole, K. and Zheng, Y. (2012)
Generation and live imaging of an endogenous Cdx2 reporter mouse line.
Genesis 50: 775-782. - McDole, K., Xiong, Y., Iglesias, P.A. and Zheng, Y. (2011)
Lineage mapping the pre- implantation mouse embryo by two-photon microscopy, new insights into the segregation of cell fates.
Dev. Biol. 355: 239-249.
Group Members
- Sonia Agüera Gonzalez
- Katharine Goodwin
- Jenny Kretzschmar
- Ewa Paluch
- Henry Westmacott

