

 Andrew Travers started his Ph.D. at the LMB under the tutelage of the late John Smith in 1964. After this he took a couple of years out as a post-doc in Jim Watson’s lab at Harvard University where he co-discovered the first of the RNA polymerase sigma factors.
Andrew Travers started his Ph.D. at the LMB under the tutelage of the late John Smith in 1964. After this he took a couple of years out as a post-doc in Jim Watson’s lab at Harvard University where he co-discovered the first of the RNA polymerase sigma factors.
He then returned to the LMB in 1970 as a staff scientist – a position he held until his retirement in 2008. At the LMB his work focussed on the organisation and three-dimensional structure of bacterial and eukaryotic chromatin with occasional forays into the roles of chromatin-associated proteins in Drosophila development.
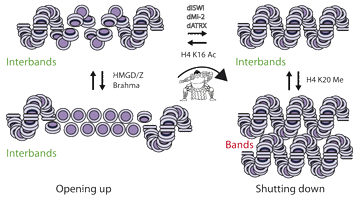
We use Drosophila genetics and biochemistry to study the mechanisms of chromatin folding and unfolding. We have recently proposed a folding pathway for chromatin which postulates that the primary role of linker histones is to increase the twist of linker DNA thus converting two connected nucleosome stacks from a twisted to a parallel configuration.
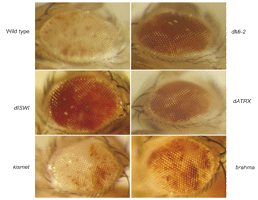
The resulting compensating writhe then facilitates the concerted packing of the parallel stacks into a 2-start helical coil. This model predicts that proteins which twist or untwist the linker DNA will respectively fold and unfold the fibre. We have shown that in Drosophila chromatin folding, as assayed by the formation of pericientric heterochromatin, requires three remodelling assemblies, dISWI, dATRX and dMi-2 as well as an active sumoylation pathway.
Conversely two other remodellers, brahma and kismet, as well as the abundant HMGB proteins (which untwist linker DNA) are without effect.

Selected Papers
- Berger, M., Farcas, A., Geertz, M., Zhelyazkova, P., Brix, K., Travers, A. and Muskhelishvili, G. (2010)
Coordination of genomic structure and transcription by the major bacterial nucleoid-associated protein HU
EMBO Rep 11: 59-64 - Sobetzko, P., Travers, A. and Muskhelishvili, G. (2012)
Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle.
Proc. Natl. Acad. Sci. USA 109: E49-E50 - Sobetzko, P., Glinkowska, M., Travers, A. and Muskhelishvili, G. (2013)
DNA thermodynamic stability and supercoil dynamics determine the gene expression program during the bacterial growth cycle.
Mol. Biosyst 9: 1643-1651 - Wu, C. and Travers, A. (2019)
Modelling and DNA topology of compact 2-start and 1-start chromatin fibres.
Nucleic Acids Res 47: 9902-9924. doi: 10.1093/nar/gkz495 - Travers, A. and Muskhelishvili, G. (2020)
Chromosomal organisation and regulation of genetic function in Escherichia coli integrates the DNA analog and digital information
EcoSal Plus 9: 10.1128/ecosalplus.ESP-0016-2019 - Travers, A. (2022)
Why DNA?
Cambridge University Press, Cambridge,