

Matteo Allegretti
The macromolecular basis of compartment remodelling
matteoall@mrc-lmb.cam.ac.ukPersonal group site
The cell is the simplest living metabolic entity. Organelles are cellular compartments specialized in a certain function. The nucleus is the essential cell compartment containing the genetic information. Our research aims to understand the molecular basis of how the nucleus is remodelled in shape during differentiation processes or due to environmental cues. Defects or anomalies during remodelling are directly involved in the biogenesis of diseases like infertility, developmental disorders and cancer metastatic progression, therefore a molecular understanding of such processes is needed to lead towards new therapeutic targets.
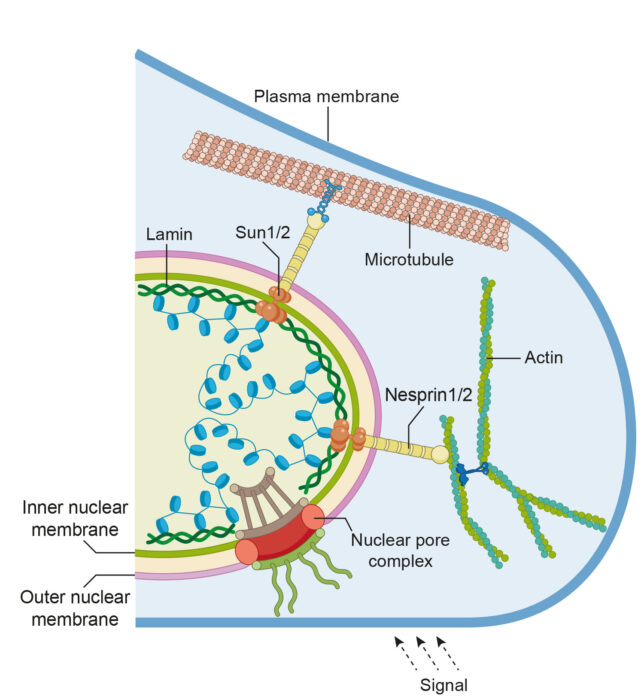
The nucleus is a very elastic organelle whose shape is regulated by the interplay between several macromolecular complexes that assemble and disassemble in response to physiological stimuli. Until now the understanding of such processes in molecular details is limited. Our lab aims to investigate the structural organization and conformational adaptations of protein complexes which shape the nucleus, and whose mutations are linked to diseases. Macromolecules include cytoskeleton and nuclear envelope proteins.
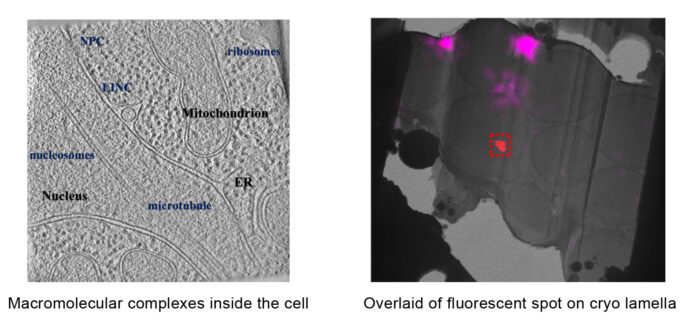
Furthermore, our lab aims to develop methods which bridge the fields of structural and cell biology, in particular to integrate electron microscopy, light microscopy and molecular modelling to investigate the structural dynamics of protein interactions in their cellular context.
Selected Papers
- Zila V., Margiotta E., Turoňová B., Müller T.G., Zimmerli C.E., Mattei S., Allegretti, M., Börner K., Rada J., Müller B., Lusic M., Kräusslich H-G., Beck M. (2021)
Cone-shaped HIV-1 capsids are transported through intact nuclear pores
Cell 184(4): 1032-1046. - Zimmerli CE, Allegretti M, Rantos V, Goetz SK, Obarska-Kosinska A, Zagoriy I, Halavatyi A, Hummer G, Mahamid J, Kosinski J, Beck M. (2021)
Nuclear pores dilate and constrict in cellulo.
Scienceeabd9776. doi: 10.1126/science.abd9776. - Allegretti, M., Zimmerli, C.E., Rantos, V., Wilfling, F., Ronchi, P., Fung, H.K.H., Lee, C-W., Hagen, W., Turoňová, B., Karius, K., Börmel, M., Zhang, X., Müller, C.W., Schwab, Y., Mahamid, J., Pfander, B,. Kosinski, J., Beck M. (2020)
In-cell architecture of the nuclear pore and snapshots of its turnover
Nature 586(7831): 796-800. - Lee, C-W., Wilfling, F., Ronchi, P., Allegretti, M., Mosalaganti, S., Jentsch S., Beck M., Pfander, B. (2020)
Selective autophagy degrades nuclear pore complexes
Nat Cell Biol. 22(2): 159-166. - Thaller, D.J., Allegretti, M., Borah, S., Ronchi, P., Beck, M., Lusk, C.P. (2019)
An ESCRT-LEM protein surveillance system is poised to directly monitor the nuclear envelope and nuclear transport system
Elife 8: e45284. - Lele , T.P., Dickinson, R.B., Gundersen, G.G. (2018)
Mechanical principles of nuclear shaping and positioning
J Cell Biol. 217(10): 3330-3342. - Cossio, P., Allegretti, M., Mayer, F., Müller, V., Vonck, J., Hummer, G. (2018)
Bayesian inference of rotor ring stoichiometry from electron microscopy images of archaeal ATP synthase
Microscopy (Oxf) 67(5): 266-273. - Elosegui-Artola, A., Andreu, I., Beedle, A.E.M., Lezamiz, A., Uroz, M., Kosmalska, A.J., Oria, R., Kechagia, J.Z., Rico-Lastres, P., Le Roux, A-L., Shanahan , C.M., Trepat, X., Navajas, D., Garcia-Manyes, S., Roca-Cusachs, P. (2017)
Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores
Cell 171(6): 1397-1410. - Bell, E.S., Lammerding J. (2016)
Causes and consequences of nuclear envelope alterations in tumour progression
Eur J Cell Biol. 95(11): 449-464. - Denais, C.M., Gilbert, R.M., Isermann, P., McGregor, A.L., te Lindert, M., Weigelin, B., Davidson, P.M., Friedl, P., Wolf, K., Lammerding J. (2016)
Nuclear envelope rupture and repair during cancer cell migration
Science 352(6283): 353-8. - Allegretti, M., Klusch, N., Mills, D.J., Vonck J., Kühlbrandt, W., Davies, K.M. (2015)
Horizontal membrane-intrinsic alpha-helices in the stator a-subunit of an F-type ATP synthase
Nature 521(7551): 237-40. - Allegretti, M., Mills, D.J., McMullan, G., Kühlbrandt, W., Vonck J. (2014)
Atomic model of the F420-reducing [NiFe] hydrogenase by electron cryo-microscopy using a direct electron detector
Elife 3: e01963. - Gundersen, G.G., Worman, H.J. (2013)
Nuclear positioning
Cell 152(6): 1376-89. - Mammoto, T., Mammoto, A., Ingber, D.E. (2013)
Mechanobiology and developmental control
Annu Rev Cell Dev Biol. 29: 27-61. - Fabian, L., Brill, J.A. (2012)
Drosophila spermiogenesis: Big things come from little packages
Spermatogenesis 2(3): 197-212.
Group Members
- Tom Dendooven
- Alia dos Santos
- Thomas Hale
- Victoria Hale
- Oliver Knowles
- Piotr Kolata
- Mart Last