

Most organisms display ~24-hour cycles in their biology. In humans and other animals, these circadian rhythms result from daily timing mechanisms in every cell that together function like a biological clock; allowing our physiology to anticipate and prepare for the differing demands of day and night. Normally our biological clock is fine-tuned each day by the schedule we keep, particularly the timing of meals and light exposure. When we see bright light or eat at the wrong biological time, as often occurs during shift work or jet-lag, it disrupts our biological clock and increases the risk of chronic illnesses such as type II diabetes, cardiovascular disease and some cancers. Conversely, the effectiveness of some drugs and surgeries can vary with the biological time of treatment. Delineating the molecular mechanisms that impart daily rhythms to our biology is therefore important for understanding human health and may provide new insights into the prevention and treatment of many diseases.
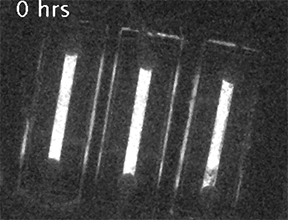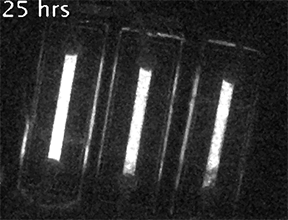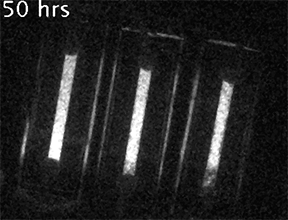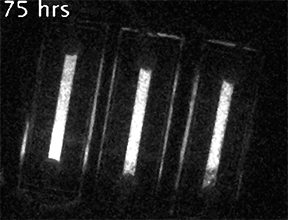

Our current research is focused on understanding the fundamental mechanisms of daily cellular timekeeping and how circadian regulation of biological function is achieved. To achieve these goals we employ a wide range of molecular biology, proteomic, metabolomic and biochemical techniques, supported by real-time fluorescent and bioluminescent reporters.
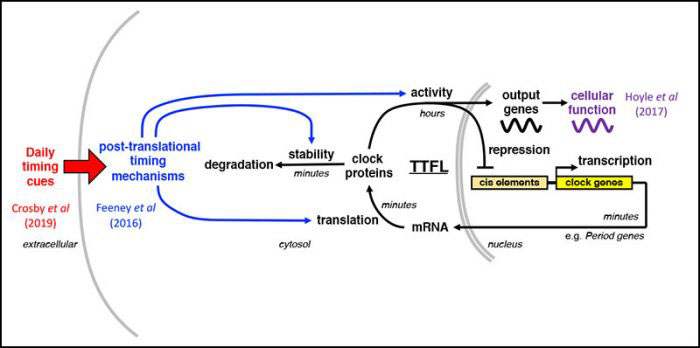
Recently we found that post-prandial increases in insulin signaling synchronise circadian rhythms throughout the body with feeding time, by increasing the production of PER clock proteins (Crosby et al 2019, Cell).
Selected Papers
- Watson et al. (2023)
Macromolecular condensation buffers intracellular water potential
Nature 623(7988): 842-852 - Beale et al. (2023)
Mechanisms and physiological function of daily haemoglobin oxidation rhythms in red blood cells
EMBO J. 42(19): - Wong et al. (2022)
CRYPTOCHROMES promote daily protein homeostasis
EMBO J 41(1): - Stangherlin, A., et al. (2021)
Compensatory ion transport buffers daily protein rhythms to regulate osmotic balance and cellular physiology
Nature Communications 12(1): 6035 - Putker M., et al. (2021)
CRYPTOCHROMES confer robustness, not rhythmicity, to circadian timekeeping
EMBO J. 40(7): e106745. - Crosby, P., et al. (2019)
Insulin/IGF-1 drives PERIOD synthesis to entrain circadian rhythms with feeding time.
Cell 177(4): 896-909. - Hoyle, N. P., et al. (2017)
Circadian actin dynamics drive rhythmic fibroblast mobilization during wound healing.
Science Translational Medicine 9(415): pii: eaal2774. - Feeney K. A., et al. (2016)
Daily magnesium fluxes regulate cellular timekeeping and energy balance
Nature 532(7599): 375-379.
Group Members
- Aymen al-Rawi
- Andrew Beale
- Anna Edmondson
- Nathan James
- Joseph Menzies
- Andrei Mihut
- Tea Stapar