 ‘Synthetic biology’ is a scientific approach that seeks to answer fundamental questions in biology by reconstruction and modification of the molecules and processes of life. Beyond its well-known role as the carrier of genetic information, DNA (and its close cousin RNA) have shown great promise as a nano-molecular building material: by careful arrangement of the bases A, T, C and G, DNA strands can be programmed to fold into specific 3D shapes. Work by Philipp Holliger’s group in the LMB’s PNAC Division showed that simple self-folding 3D nano-scale objects can be constructed from materials other than DNA and RNA, specifically from a series of ‘synthetic genetic polymers’, also known as xeno nucleic acids (XNAs). XNAs are composed of strings of chemical building blocks that do not occur in nature, and display a much wider range of physicochemical properties than DNA, including much higher biostability. This is the first time such ‘DNA nanotechnology’ designs have been built entirely from unnatural chemical building blocks.
‘Synthetic biology’ is a scientific approach that seeks to answer fundamental questions in biology by reconstruction and modification of the molecules and processes of life. Beyond its well-known role as the carrier of genetic information, DNA (and its close cousin RNA) have shown great promise as a nano-molecular building material: by careful arrangement of the bases A, T, C and G, DNA strands can be programmed to fold into specific 3D shapes. Work by Philipp Holliger’s group in the LMB’s PNAC Division showed that simple self-folding 3D nano-scale objects can be constructed from materials other than DNA and RNA, specifically from a series of ‘synthetic genetic polymers’, also known as xeno nucleic acids (XNAs). XNAs are composed of strings of chemical building blocks that do not occur in nature, and display a much wider range of physicochemical properties than DNA, including much higher biostability. This is the first time such ‘DNA nanotechnology’ designs have been built entirely from unnatural chemical building blocks.
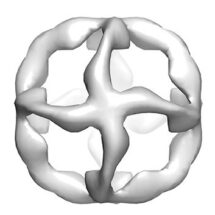
Alex Taylor and colleagues at the LMB, together with collaborators at The Institute of Cancer Research, London, and KU Leuven, Belgium used polymerase enzymes previously engineered in Philipp’s lab to synthesise XNA strands designed to bind to one another and fold into simple geometric designs (a tetrahedron or an octahedron) following previous designs for DNA nanotechnology. Although distinct from DNA and RNA, the XNAs still carry the bases A, T, C and G, so the standard Watson-Crick base pairing rules still operate to ‘program’ XNA strands to bind to each other in a defined manner. The XNA nanostructures were visualised using electron microscopy to demonstrate the structures formed. The chemical uniformity of DNA and its susceptibility to DNA-cutting enzymes is a well-known problem – DNA is degraded in blood serum within hours (and RNA within minutes). This work demonstrates that non-natural XNA chemistries offer novel building materials for the construction of 3D nanostructures that resist degradation in serum, and opening the door for the development of designs with properties beyond those of DNA and RNA.
Programmable ‘DNA nanotechnology’ has many potential applications in research, industry and medicine. In principle, nanostructures capable of sensing and responding to disease-associated molecules (e.g. molecules on the surface of tumour cells) could be used as the basis of targeted drug delivery. XNA-based designs could substantially improve the efficacy of such strategies, however the unusual structures of XNAs mean that their use in more complicated architecture may require a more detailed investigation into XNA self-association rules.
The work was funded by the MRC, the European Science Foundation, the Biotechnology and Biological Sciences Research Council and Cancer Research UK.
Further references:
Paper in ChemBioChem
Philipp’s group page
Edward Morris’ group page
Piet Herdewijn’s group page New Scientist article