Mechanism-based, light-activated traps allow protease and hydrolase substrate discovery in cells
Hydrolase enzymes, including proteases, are vital for catalysing the synthesis and fragmentation of molecules in the cell. These enzymes are encoded by two to three per cent of genes in the human genome and fourteen per cent of these enzymes are active drug targets. Despite this, the substrates and activities of many hydrolases largely remain unknown. Now, Jason Chin’s group in the LMB’s PNAC Division have designed a new technique to identify hydrolase substrates in complex mixtures and in live mammalian cells.
The new method builds on previous work from Jason’s lab in which genetic code expansion is used to replace the catalytic serine or cysteine in a hydrolase with the amine containing analog, 2,3-diaminopropionic acid (Dap). The resulting hydrolase undergoes the first step reaction with substrates to covalently link a substrate fragment to the enzyme. Shan Tang, in Jason’s group, developed the new approach – in which hydrolases containing Dap are unveiled by a pulse of light in live mammalian cells – and showed that this allowed the trapping and identification of protease and hydrolase substrate in live cells. Using her approach, Shan discovered the activity of an orphan hydrolase (RBBP9). She then collaborated with Matthew Freeman’s group at the Sir William Dunn School of Pathology, University of Oxford to address the outstanding challenge of discovering substrates for an intramembrane protease (RHBDL4).
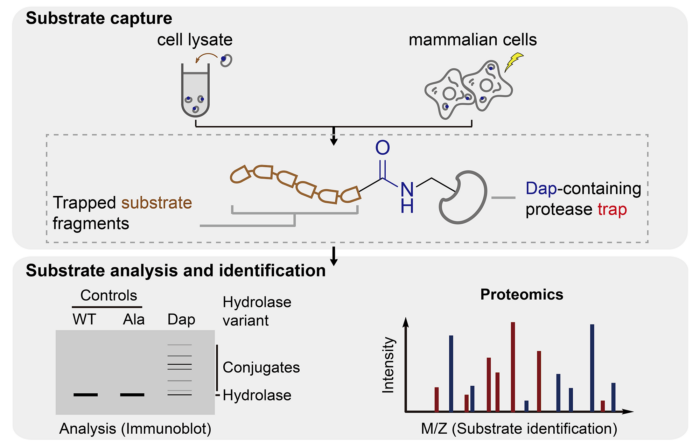
This research highlights a new exemplar strategy for the discovery of new substrates and activities of hydrolase enzymes. The research holds clinical significance, as the abnormal regulation of hydrolase activity is a key feature of numerous diseases.
This work was funded by UKRI MRC, The Wellcome Trust, EMBO and The Marie Skłodowska-Curie Actions.
Further references
Mechanism-based traps enable protease and hydrolase substrate discovery. Tang, S., Beattie, AT., Kafkova, L., Petris, G., Huguenin-Dezot, N., Fiedler, M., Freeman, M., Chin, JW. Nature
Jason’s group page
Matthew Freeman – Sir William Dunn School of Pathology
Previous Insight on Research article
Catching enzymes in the act of making an antibiotic
Cells reprogrammed for genetically encoded polymer synthesis and viral resistance
Creating an entire bacterial genome with a compressed genetic code