The receptor TECPR1 detects sphingomyelin exposed on damaged membranes in the cytosol and conjugates the ubiquitin-like autophagy protein LC3 to those membranes
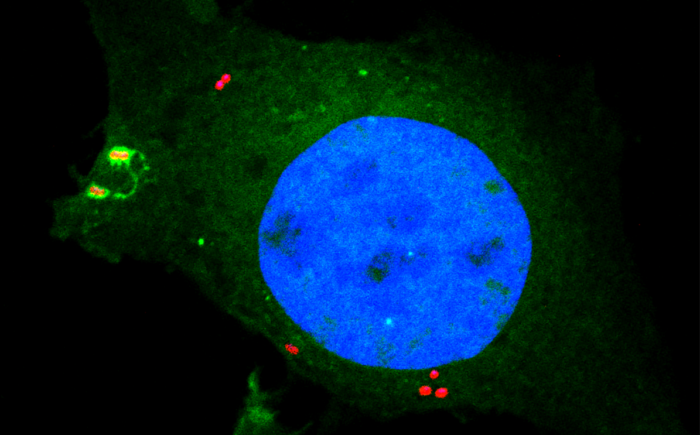
Cellular homeostasis is reliant upon membrane integrity, and therefore the speedy repair or disposal of damaged membranes is crucial to cellular health. Thus, cells rely on the immediate and receptive detection of any surface disturbance. Alongside monitoring their plasma membranes, cells use damage to internal membranes as a warning sign indicating pathogen invasion, for example when pathogens enter the host cytosol via an endosomal or phagosomal compartment.
Bacteria containing vacuoles (BCVs) chaperone invasive bacteria into host cells and subsequent rupture of BCVs allows these bacteria to enter to cytosol. This process exposes otherwise shielded danger signals such as glycans and sphingomyelin. It has been established that glycans are detected by galectin-8, engagement of which then triggers anti-bacterial autophagy, but how cells sense and respond the cytosolically-exposed sphingomyelin is unclear. Felix Randow’s group, in the LMB’s PNAC Division, has identified TECPR1 (Tectonin Beta-Propeller Repeat Containing 1) as a receptor for cytosolically exposed sphingomyelin.
Keith Boyle, a member of the group, used sphingomyelin-containing liposomes to enrich binding proteins from cell lysates in order to identify TECPR1 as a sphingomyelin-binding protein. Further experimentation by Sascha Martens, a former LMB member and now a professor at the University of Vienna, revealed that TECPR1 recruits Autophagy-Related Gene 5 (ATG5) into a new E3 ligase complex that conjugates the ubiquitin-like protein LC3 to membranes – a process essential for the execution of autophagy. While there are hundreds of E3 ligases that conjugate ubiquitin onto substrates, the sphingomyelin-activated TECPR1-ATG5 complex is only the second ligase known to act on LC3.
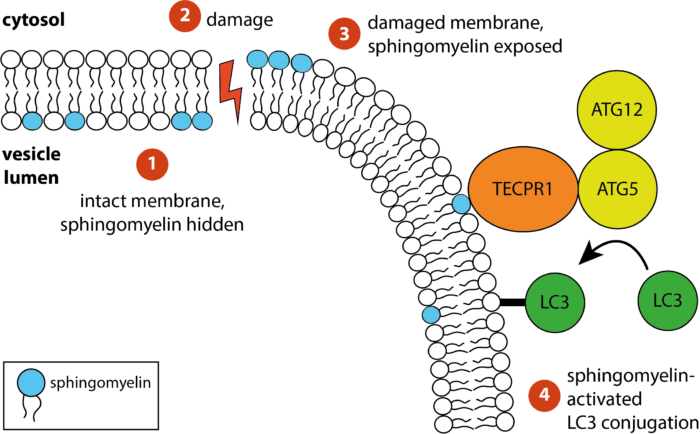
Bacteria-containing vacuoles with cytosolically exposed sphingomyelin will ultimately burst and release their content into the nutrient-rich cytosol – thereby enabling rapid bacterial proliferation to the detriment of the cell and eventually the host organism. Though burst vesicles are detected owing to the exposure of glycans, detection of sphingomyelin via TECPR1 before they burst could buy the cell extra time to prepare for imminent bacterial entry. The group posits that cells deploy TECPR1 to induce non-canonical autophagy by labelling stressed, sphingomyelin-positive intracellular membranes with LC3.
Furthermore, the group identified that TECPR1 binds sphingomyelin through its N-terminal DysF domain. Assisted by Paul Elliott, another former LMB member and now a Group Leader at the University of Oxford, the group used X-ray crystallography to solve the structure of the DysF domain. This is the first domain identified to detect membrane stress through the binding of sphingomyelin.
Alongside its contribution to our understanding of cells, this research holds further significance as the canonical autophagy pathway is considered a promising drug target for metabolic intervention, for example in cases of obesity or diabetes. TECPR1 represents an alternative drug target into the autophagy process. Furthermore, as TECPR1 detects general pathogen-induced host cell damage, rather than specific pathogens directly, it is predicted to sense a wide variety of pathogens with numerous implications for human health.
This work was funded by UKRI MRC and the Wellcome Trust.
Further references
TECPR1 conjugates LC3 to damaged endomembranes upon detection of sphingomyelin exposure. Boyle, KB., Ellison, CJ., Elliott, PR., Schuschnig, M., Grimes, K., Dionne, MS., Sasakawa, C., Munro, S., Martens, S., Randow, F. The EMBO Journal.
Felix’s group page
Previous Insight on Research articles
A new way to mark bacterial invaders for destruction
Cell-invading bacteria are converted into signalling platforms that may trigger septic shock