Use of full length eIF2 substrate and combination of approaches reveals substrate recruitment mechanism of phosphatase non-catalytic subunit, PPP1R15B
The reversible phosphorylation of proteins controls virtually all aspects of cell and organism function through the opposing actions of kinases and phosphatases. Mechanistic understanding of these enzymes has the potential to open new therapeutic opportunities. Now, for the first time, researchers in Anne Bertolotti’s group in the LMB Neurobiology Division have overcome two key challenges to reveal how a major phosphatase non-catalytic subunit, PPP1R15B, binds to its full 125 kDa trimeric protein substrate, eukaryotic translation initiation factor 2 (eIF2).
PP1 is a major serine/threonine protein phosphatase which exists in complex with hundreds of non-catalytic subunits forming a wide repertoire of holophosphatases. One of the known substrates of a PP1 holophosphatase is eIF2. Phosphorylation of eIF2 alpha subunit is a vital stress response signalling event which is regulated by four kinases and two holophosphatases, PP1 bound to PPP1R15A (R15A) or PPP1R15B (R15B). Because R15B is constitutively expressed, whereas R15A is stress-induced, the Bertolotti team set out to understand how R15B binds to eIF2.
Whilst the mechanism of PP1 recruitment to its non-catalytic subunits is well understood, how substrates are recruited to PP1 holoenzymes has remained largely unknown for at least two reasons. The non-catalytic subunits of the PP1-containing holophosphatases, including R15B, are often intrinsically disordered, posing an enormous challenge for structural biology. In addition, the substrates are often large proteins or complexes which are difficult to purify and study. Because eIF2 is a large trimeric complex of 1,120 amino acids, previous studies have only used a smaller approximately 180 amino-acid N-terminal fragment.
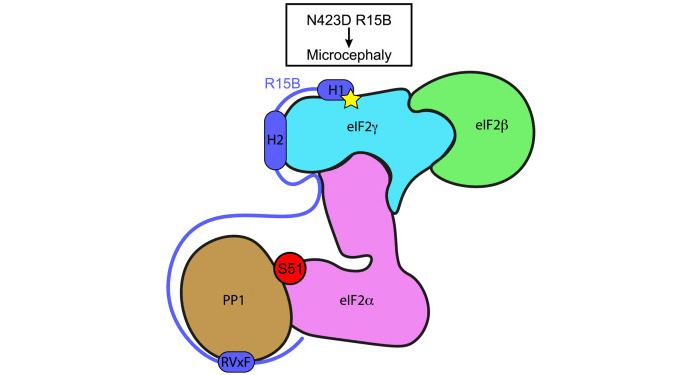
To overcome the challenges posed by full-length eIF2 and intrinsically disordered R15B, the Bertolotti group used a combination of approaches consisting of structural proteomics, nuclear magnetic resonance (NMR) and functional analyses. Hydrogen/deuterium exchange mass spectrometry (HDX-MS) analyses, conducted in collaboration with the LMB Mass Spectrometry Facility, revealed that R15B binds to the far end of the eIF2 complex relative to the N-terminal phosphorylation site. This was a surprise because prior studies assumed the binding occurred near the phosphorylated residue. NMR analyses, carried out together with the LMB NMR Facility, revealed that R15B is largely disordered, except for three short helical regions, H1, H2 and H3. These helical regions were also predicted by Alphafold2. Further NMR and mass spectrometry analysis showed R15B binds to eIF2 via H1 and H2, and to a lesser extent H3.
At the same time, collaborators Marjon van Slegtenhorst and Laura Donker Kaat, from the Department of Clinical Genetics at Erasmus MC in Rotterdam, identified a homozygous missense variant in a residue of R15B adjacent to H1 in a patient with microcephaly, developmental delay and intellectual disability. Further investigation by the Bertolotti group found that the mutation impaired R15B substrate binding and dephosphorylation, establishing the molecular mechanism of this rare condition.
Taken together, the team’s experimental findings explain how a large substrate can be captured by short helical elements within an intrinsically disordered protein to efficiently promote its dephosphorylation. The key to revealing this phosphatase recruitment mechanism was the use of its full 125 kDa protein substrate, highlighting the importance of using full length substrate in studies of other PP1 holoenzymes.
This work was funded by UKRI MRC, the European Research Council, a Wellcome Trust Principal Investigator Award, Max Planck Institute for Multidisciplinary Sciences and the Max Planck Society.
Further references
Recruitment of trimeric eIF2 by phosphatase non-catalytic subunit PPP1R15B. Fatalska, A., Hodgson, G., Freund, S.M.V., Maslen, S.L., Morgan, T., Thorkelsson, S.R., van Slegtenhorst, M., Lorenz, S., Andreeva, A., Donker Kaat, L., and Bertolotti, A. Molecular Cell
Anne’s group page
Department of Clinical Genetics at Erasmus MC in Rotterdam
Previous Insight on Research articles
Activation of the integrated stress response by inhibitors of its kinases
Making the undruggable druggable: the first platform to discover selective phosphatase inhibitors
Uncovering the action of a selective holophosphatase inhibitor