Discovery of a novel pathway to rescue stalled mitoribosomes and structural characterisation of the rescue factors
Mitochondria are descendants of bacteria that were engulfed by a larger archaeal cell about 2 billion years ago. Most of their original bacterial genes have been lost or transferred to the host cell, but they still maintain a small genome that makes several proteins essential for their function. These include enzymes involved in the production of the energy storage molecule ATP through a process known as oxidative phosphorylation where molecular oxygen is used for respiration.
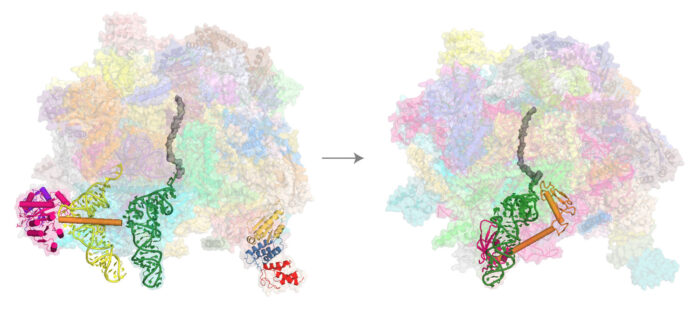
To translate their own genes, mitochondria have their own ribosomes, known as mitoribosomes. Mitoribosomes are quite different from those in the cytoplasm, and little is known about their quality control mechanisms, including whether any pathway exists to rescue stalled mitoribosomes. Venki Ramakrishnan’s group, in the LMB’s Structural Studies Division, has discovered a novel rescue pathway and determined the structures of the responsible rescue factors in action.
Defective translation can result in the ribosomes stalling and consequently, mitochondrial impairment. A rescue mechanism would allow stalled ribosomes and associated translational factors to be recycled for future use, as well as degrade the incomplete polypeptide, thus preventing accumulation of misfolded proteins. However, no such rescue mechanism has previously been found for mitoribosomes.
Nirupa Desai, Hanting Yang, and Vish Chandrasekaran, in Venki’s group, in collaboration with Michal Minczuk at the MRC Mitochondrial Biology Unit, obtained cryo-EM structures of mitoribosomes purified from human cell lines known to have a high propensity for ribosome stalling. Exhaustive analysis of the cryo-EM datasets allowed them to capture structures of most of the steps in the mitoribosomal elongation cycle as well as uncover a novel rescue pathway involving a release factor they named mtRF-R and its binding partner, an RNA binding protein, MTRES1. Their structures show that these rescue factors function on the large mitoribosomal subunit of stalled and split ribosomes, to eject the nascent polypeptide chain and release the peptidyl tRNA, something that they confirmed with assistance from Razina Kazi, also in Venki’s group.
This work sets the stage for further understanding of the regulation of translation in mitochondria. A detailed comparison of ribosomal function in mitochondria versus bacteria would benefit our understanding of why some antibiotics might have unwanted effects on the function of our own cells. Furthermore, mutations in mtRF-R have been associated with neurological disorders, including optic atrophy, peripheral neuropathy, and spastic paraparesis. The discovery of its role in mitoribosomal rescue can help provide a molecular understanding of why mutations in this protein cause disease.
The work was funded by UKRI MRC, Wellcome Trust, the Louis-Jeantet Foundation, the Agouron Institute, and EMBO.
Further references
Elongational stalling activates mitoribosome-associated quality control. Desai, N., Yang, H., Chandrasekaran, V., Kazi, R., Minczuk M., Ramakrishnan, V. Science, 370(6520): 1105-1110.
Venki’s group page
Michal Minczuk’s group page
Previous Insights on Research
Breakthrough in structural biology reveals mitochondrial ribosome structure
The idiosyncratic ribosomes of mitochondria