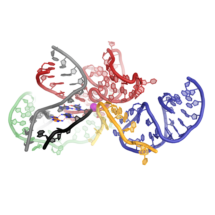
The spliceosome is a molecular machine that plays an important role in gene expression. It cuts non-coding sequences (introns) out of messenger RNA (mRNA) precursors, and stitches together the useful coding sequences (exons). The spliceosome performs this in two steps. First, the start of an intron is recognised, cut, and joined to a specific point in the middle of that intron, forming a lasso-like looped structure. In the second step, the end of the intron is recognised, cut, and the exons joined together. Previous research by Kiyoshi Nagai’s group in the LMB’s Structural Studies Division has shown how the spliceosome assembles and performs the first step, and how the spliceosome is remodelled for the second step. Now, after decades of research by many scientists, Kiyoshi’s group has revealed how the spliceosome performs the second step and recognises the end of the intron, completing the puzzle of splicing catalysis.
Max Wilkinson, with the help of Sebastian Fica and other members of the group, purified spliceosomes from baker’s yeast, mutated so the spliced mRNA could not be released. Using cryo-electron microscopy they then solved the structure of the spliceosome immediately after it had completed the second step of splicing. This revealed that, remarkably, the end of the intron is recognised directly by the branched looped intron structure formed after the first step. The very last base of the intron forms an unorthodox base pair with the very first base of the intron. The penultimate base of the intron forms a different, but also unorthodox base pair with the branch point in the intron. Thus, the two steps of splicing are closely related: completion of the first step forms the structure required for the second. These results explain why the introns of genes start with GU and end with AG.
Until now, all substrate sequence recognition by the spliceosome has been via base pairing to the special snRNAs that form the catalytic centre of the spliceosome. Thus the mechanism revealed by this new research – whereby the end of the intron is recognised by the intron itself – was totally unexpected.
It is estimated that 15-30% of human genetic diseases involve defects in splicing. Diseases can arise when sequences in the messenger RNA are mutated and can no longer be recognised by the spliceosome machinery. It is therefore crucial to understand the mechanism by which splice sites are selected and this research sheds important new light on the process.
This research was funded by the Medical Research Council, European Research Council, a Cambridge-Rutherford Memorial PhD scholarship and a Marie-Curie fellowship.
Further references:
Paper in Science
Kiyoshi’s group page
Previous Insight on Research: New insights into the structure and dynamics of the catalytic spliceosome
Previous Insight on Research: Structure of the catalytic spliceosome
Previous Insight on Research: Cryo-EM sheds new light on the spliceosome activation process