Revealing the structure of the drug favipiravir bound to the SARS-CoV-2 RNA polymerase could benefit efforts to develop new COVID-19 treatments that block viral replication
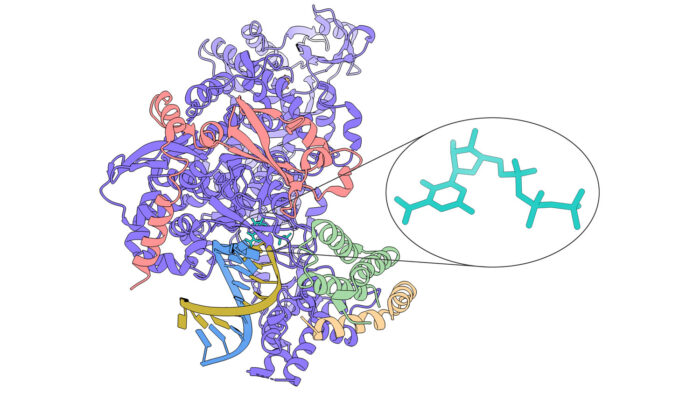
One promising drug target for treatment of COVID-19 is the enzyme responsible for copying the SARS-CoV-2 viral RNA, as it is crucial for replication of the virus. Several small molecule drugs that have a broad spectrum of activity against other viral polymerases are being trialled against SARS-CoV-2, but the failure of some of these demonstrates the importance of a better understanding of how they work, so that they can be improved upon. The structure of one such molecule, favipiravir, in complex with the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp), has now been solved by a collaboration between Chris Russo’s, David Barford’s, Jan Löwe’s, and John Sutherland’s groups at the LMB, providing new insight into its mechanism of action.
Favipiravir is a small molecule with a similar structure to the bases of the purines guanosine and adenosine. In cells favipiravir is converted into favipiravir ribosyl triphosphate (favipiravir-RTP) that mimics GTP, allowing it to interact with the polymerase enzyme that normally adds nucleotides to a growing RNA chain during RNA replication. Favipiravir is currently in clinical trials as a treatment for SARS-CoV-2 infection, but little is known about its precise molecular mechanism of action. Remdesivir is another nucleoside analogue that was trialled against SARS-CoV-2, but was found to have limited beneficial effect but is still being used in some cases. Structural insights into how favipiravir might inhibit viral replication would be of great benefit for the design of small molecules that function as more effective treatments for COVID-19.
To investigate this, Katerina Naydenova, Kyle Muir, and Long-Fei Wu performed RNA polymerisation assays and determined the structure of favipiravir-RTP in complex with the SARS-CoV-2 RdRp using cryo-EM. The polymerisation assays indicated that favipiravir-RTP is only weakly incorporated into a new RNA strand, but nonetheless suppresses RNA replication by acting as a competitive inhibitor of GTP. The structure revealed that favipiravir-RTP is bound to the catalytic site of RdRp in an unusual conformation, such that the part of the molecule that should covalently bond with the primer RNA chain is not in the correct place, explaining the low rate of incorporation into the RNA strand.
It had been thought that a possible mechanism of favipiravir against SARS-CoV-2 would be that incorporation into new RNA chains would lead to increased rates of lethal mutagenesis, but this study suggests that its rate of incorporation might be insufficient to cause this. Conversely, binding to RdRp in its non-productive conformation could make favipiravir a good competitive inhibitor as it slows the rate of RNA replication by stopping natural nucleotides interacting with the polymerase. The structural insights of this work suggest ways in which new small molecules could be developed that might bind more tightly into the polymerase active site to more effectively stall RNA synthesis or that could allow greater rates of incorporation which could drive more efficient lethal mutagenesis. Either of these are important potential directions to produce new COVID-19 treatments.
The work was funded by UKRI MRC, a Vice-Chancellor’s Award from the Cambridge Commonwealth, European and International Trust, a Bradfield scholarship, and Cancer Research UK.
Further references
Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in presence of favipiravir-RTP. Naydenova, K., Muir, KW., Wu, L-F., Zhang, Z., Coscia, F., Peet, MJ., Castro-Hartmann, P., Qian, P., Sader, K., Dent, K., Kimanius, D., Sutherland, JD., Löwe, J., Barford, D., Russo, CJ. PNAS 118(7) e2021946118
Chris’ group page
David’s group page
Jan’s group page
John’s group page
LMB joins the fight against COVID-19