A comprehensive model of how evolution has driven complexity in an initiator of transcription through diverse lifeforms, from single-celled archaea to humans
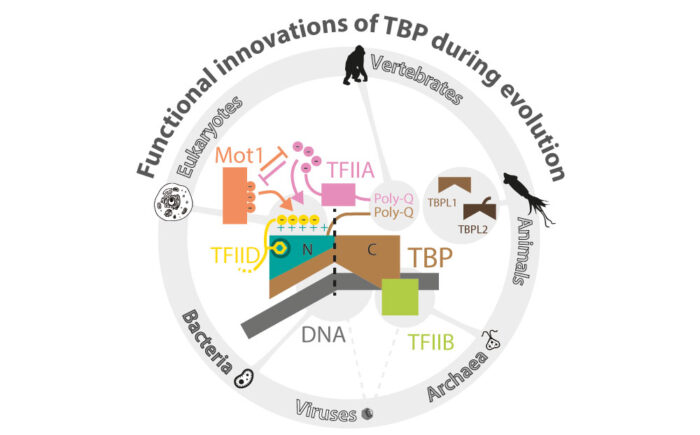
Transcription is a fundamental biological process in our cells – it is the first step in the central dogma of molecular biology by which information in our genes is converted into proteins. Transcription initiation is the primary step in this process, wherein a protein known as TBP (TATA box binding protein) plays a critical role. TBP performs this function in as diverse lifeforms as single-celled archaea and complex animals, including humans.
This work, led by Balaji Santhanam and Charles Ravarani in M Madan Babu’s group in Structural Studies Division, traced the functional evolution of TBP all the way from viruses and unicellular species to multicellular organisms. Their study provides novel insights on TBP’s molecular journey across this evolutionary time-scale and is the first comprehensive model for explaining how TBP acquired its extant functions (its “functional innovations”).
What does TBP do and why is it important?
TBP recognises and binds to specific DNA sequences (TATA boxes), and bends the DNA. This leads to the recruitment of other factors, such as adapter molecules and the RNA polymerase complex to assemble the pre-initiation complex (PIC) for mediating transcription. TBP is the critical component even in the simplest form of the PIC as seen in single-celled archaea.
Charles, Balaji and other members in Madan’s group performed a comprehensive study of the evolutionary history of TBP and its interaction partners across all kingdoms of life, including viruses.They sought to identify key conserved molecular features of TBP that are functionally relevant, and also investigate how it is exploited by their its partners.
Functional evolution of TBP
Towards this, they systematically compared the structures and sequences of TBP in organisms that emerged at different points in evolution. Their approach simultaneously considered spatial (i.e. every residue on TBP) and temporal (i.e. different points in organismal evolution) aspects of TBP. This spatio-temporal approach uncovered how concerted chemical changes on the molecule have all contributed to the functional innovations of TBP. These changes on the TBP range from a key set of amino acid residues to longer stretches of amino acids and major transitions in the form of gene duplicates.
Apart from insights on TBP’s functional evolution, the study also identified novel TBP-like proteins in poxviruses that are poorly characterised and revealed the existence of complete orthogonal TBP-adapter systems in many double-stranded DNA viruses. The research proposes how these viruses might escape their host regulatory control on transcription initiation. The orthogonality of the viral TBP systems potentially provides a novel scope for drug development targeting viral TBP without affecting the host TBP system.
This work was funded by the MRC, a European Research Council Consolidator Grant and a Lister Institute Prize fellowship.
Further references
Molecular determinants underlying functional innovations of TBP and their impact on transcription initiation. Ravarani, CNJ., Flock, T., Chavali, S., Anandapadamanaban, M., Babu, MM., Santhanam, B. Nature Communications (Epub ahead of print)
Madan’s group page
Previous Insights on Research
Understanding noise: the molecular determinants of random variation in gene expression levels
