The most complete structure of the replisome to date, providing insight into how DNA is unwound before replication and how several key factors associate during replication
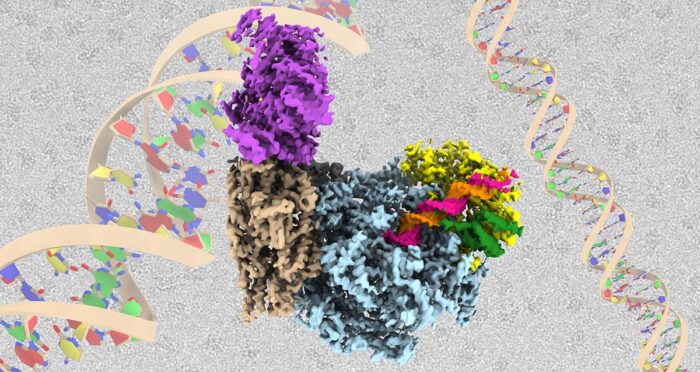
Every time a cell divides, all of its genetic material must be duplicated so that it can be passed in its entirety to both daughter cells. Accurate replication is essential to ensure normal cellular function and to prevent mutations from arising that can ultimately lead to cancer. This process is controlled by a molecular machine known as the replisome. Joe Yeeles’ group, in the LMB’s PNAC Division, has now produced the highest resolution and most complete structure of a eukaryotic replisome to date.
Two main functions are required during DNA replication: unwinding the two strands of the double helix so that each single strand can be accessed, and using those single strands as templates to reproduce new double-stranded DNA. These processes are tightly coupled by the replisome complex to avoid stretches of exposed single-stranded DNA between the point of unwinding and the point of synthesis of new DNA.
Progression of the replisome as it fulfils both of these main functions can be impeded by multiple different types of obstacles that affect either unwinding or DNA synthesis. These obstacles include proteins attached to the double-stranded DNA that block forward movement of the replisome as it untwists the DNA as well as damage to the DNA template that inhibits DNA synthesis enzymes.
Consequently, the replisome has evolved many strategies to respond to obstacles. In their two most recent papers, Joe’s group has examined the structure of the protein complex associated with unwinding of the DNA, in addition to uncovering a mechanism by which the replisome overcomes damage to the DNA template.
To better understand control of DNA unwinding, Domagoj Baretić and Michael Jenkyn-Bedford, two researchers in Joe’s group, isolated individual replisome subunits and built the complex from the ground up on a section of DNA in a test tube, before solving the structure using electron cryo-microscopy (cryo-EM) and mass spectrometry. This replisome complex consisted of the CMG helicase enzyme complex that is responsible for untwisting the helix, bound to the Fork Protection Complex that both stabilises the replisome when it stalls and aids normal replication rates. Importantly, this is the first structure of the Fork Protection Complex in the context of a replisome.
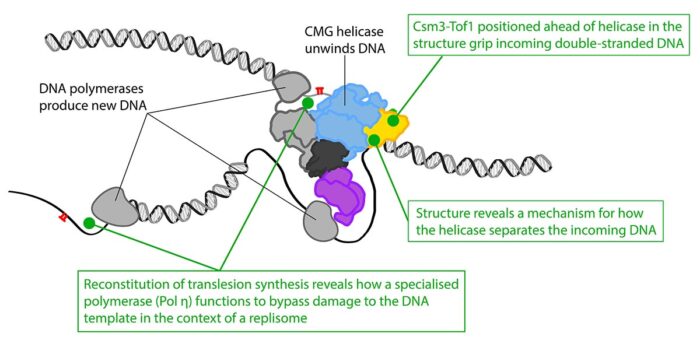
With this structure, the team could see how two of the proteins that make up the Fork Protection Complex in yeast, Csm3 and Tof1, are positioned right at the front of the replisome where they “grip” the double-stranded DNA before it is separated. This ensures that the replisome responds correctly when it encounters protein barriers on the template. The structure also reveals how the CMG helicase unwinds the DNA, as the team identified a dedicated “strand separation pin” that functions as a wedge to physically split the two strands apart as one strand is then pulled through a tunnel within CMG.
Alongside this work, Thomas Guilliam, another researcher in Joe’s group, has assembled the replisome from individual components in order to investigate how this machine overcomes damage to the DNA template rather than stalling the whole process. This was the first time the mechanism of damage tolerance known as translesion synthesis had been reconstituted with purified proteins in the context of a replisome. This allowed Thomas to show how, once damage is encountered, a different DNA synthesising enzyme that is able to tolerate the damage is recruited and how this is regulated so that the normal enzyme takes over again after the damage has been bypassed.
The replisome must copy vast quantities of DNA and must do so efficiently and accurately, whilst overcoming a large number of different types of obstacles. Deciphering its function is therefore critical to understand how cells can minimise the number of potentially cancer-causing errors. While this work presents the most complete 3D picture of the replisome to date and provides insights into how it can tolerate some forms of DNA damage, the replisome is a large molecular machine with many parts and much work remains to be done to fully understand the processes of DNA replication and repair of DNA damage. This work provides an ideal platform to build ever-more complex replisome assemblies that could lead to a complete mechanistic description of the machine.
The work was funded by UKRI MRC, the Medical Research Foundation, and Wellcome Trust.
Further references
Cryo-EM structure of the Fork Protection Complex bound to CMG at a replication fork. Baretić, D., Jenkyn-Bedford, M., Aria, V., Cannone, G., Skehel, M., Yeeles, JTP. Molecular Cell 78:1-15.
Reconstitution of translesion synthesis reveals a mechanism of eukaryotic DNA replication restart. Guilliam, TA., Yeeles, JTP. Nature Structural and Molecular Biology (Epub ahead of print)
Joe’s group page