
With the ‘resolution revolution’ of recent years, it should in principle be possible to determine atomic resolution structures of any proteins using electron cryo-microscopy (cryo-EM). However, in practice, preparation of frozen samples that are suitable for high resolution imaging is a barrier to progress. Christopher Russo’s group in the LMB’s Structural Studies Division have now developed a new cryo-EM support involving modified graphene which improves image quality, allows high resolution structure determination, and has the potential to turn sample preparation from a trial-and-error art into a reproducible scientific process.
During sample preparation, purified proteins are suspended in a very thin layer of water on a grid, which is rapidly frozen by plunging it into liquid ethane. One problem with this system is that the protein samples in the very thin layer of water can get exposed to the air-liquid interface, with detrimental effects to the structure. Covering the grid with a thin film that provides an adherent surface for the protein can reduce this risk.
A single layer of graphene, a hexagonal lattice of carbon atoms, is only one atom thick, which makes it effectively invisible in cryo-EM, and could be a near perfect support film, but a single type of adherent surface will not be sufficient for all possible protein specimens. To address this problem, Katerina Naydenova and Mathew Peet in Christopher’s group developed a method to introduce different chemical modifications to a sheet of graphene using low energy helium plasma with vapours of small molecules that will react with and modify the graphene in different ways. As well as making the graphene film suitable for cryo-EM, these modifications are able to interact with the surface of proteins in different ways and reduce movement of the sample, thereby improving image quality.
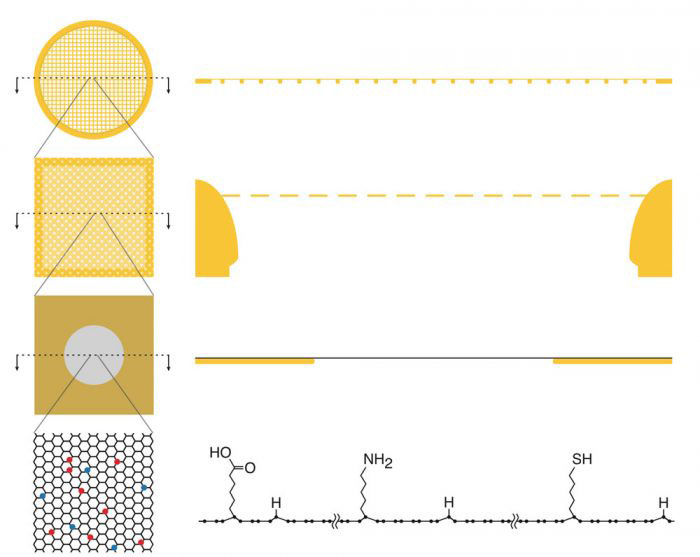
Katerina and Mathew tested this system with four different modifying small molecules and used the newly developed support for determination of the structure of the 30S ribosomal subunit. Interestingly, these experiments showed how some sides of the protein complex interacted preferentially with different chemical modifications, due to different properties in the exposed amino acids on the surface of the subunit. This shows how prior knowledge of how a protein interacts with other well-defined surfaces could help guide the choice of useful chemical graphene modifications to improve sample preparation and benefit high resolution structure determination.
Although only a small number of different modifications were tested here, it should be possible to perform this method with a wide range of small molecules. This will make it possible to have cryo-EM supports with tuneable surface properties designed for specific proteins. The team were also able to have different modifications in different regions of the support grid, which will allow scientists to test a defined set of different modifications in a reproducible way so that sample preparation will no longer be trial-and-error and high resolution structures will be easier to obtain.
The work was funded by the MRC, the Cambridge Commonwealth, European and International Trust, a Trinity College Sir John Bradfield bursary, and the Leverhulme Trust.
Further references
Multifunctional graphene supports for electron cryomicroscopy. Naydenova, K., Peet, MJ., Russo, CJ. PNAS 116(24): 11718-11724
Christopher’s group page
Previous Insight on Research: Golden grids for electron cryo-microscopy