Discovery of a novel group of optimal reverse transcriptase enzymes for RNA and XNAs that could aid development of diagnostics and therapeutics for many diseases
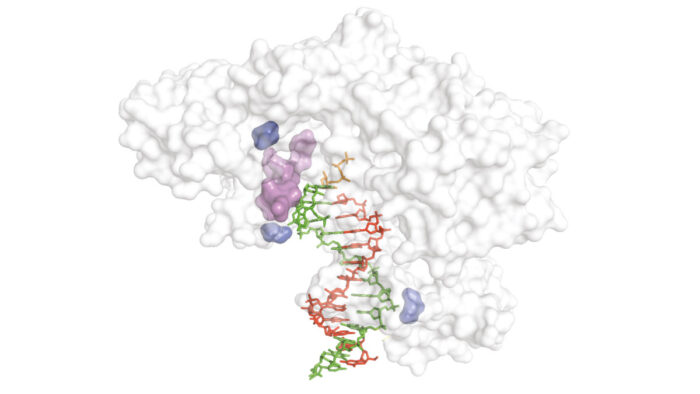
As well as being required for some of the core techniques of molecular biology, reverse transcriptase (RT) enzymes have played a key role in synthetic genetics by enabling synthesis, replication, and evolution of xeno-nucleic acids (XNAs). However, for most XNA chemistries, no RT enzymes are available or existing enzymes have low activity. Philipp Holliger’s group, in the LMB’s PNAC Division, have developed a new directed evolution method to improve RT activity for any nucleic acid chemistry and discovered a new group of optimal RT enzymes.
XNAs are genetic polymers like DNA or RNA, but with altered sugar rings, bases, or backbones. Despite these differing chemistries, they are still able to store and pass on genetic information and can perform enzymatic functions much like RNA enzymes, also known as ribozymes. They can also function as aptamers and bind to proteins with high specificity and affinity, just as antibodies do. These functions and the varied properties resulting from their different chemistries mean that XNAs could have a wide range of applications in biotechnology and medicine. However, generation of XNA aptamers and XNAzymes has been limited by lack of high-fidelity RT enzymes.
What is reverse transcription?
The first step of the so-called central dogma of molecular biology is the transcription of DNA to produce RNA. RNA can also be reverse transcribed to produce DNA and synthetic RT enzymes allow access to XNAs.
By enabling scientists to convert RNA to DNA, RT enzymes make it possible for scientists to more easily study what genes are being transcribed inside cells, and therefore which genes are “on”, through core techniques like RT-PCR and RNAseq. Alongside applications in research, this capacity is also used for medical tests, such as to test for the presence of viral RNA, including in COVID-19 tests.
To address the lack of high-fidelity RT enzymes, Gillian Houlihan and others from Philipp’s group developed a new directed evolution technique that led to the discovery of a new group of optimal RT enzymes that can decode genetic information more accurately and more efficiently. Importantly, this new method is compatible with any nucleic acid chemistry and their discovery includes novel RT enzymes for XNA chemistries for which no RT enzyme previously existed. Amongst the new RT enzymes are the first enzymes capable of actively proof-reading during XNA reverse transcription, improving accuracy.
High fidelity RNA RT enzymes will have immediate applications in research and biotechnology as they will provide improved sequencing accuracy when analysing cellular or viral RNAs. Improved XNA RT activity will likely aid development of novel XNA aptamers that could be useful in diagnostics and therapeutics for a wide range of diseases. As a specific example, this work includes the first RT enzyme for the XNA chemistry used in the anti-sense oligo drug nusinersen that has FDA and EMA approval for treatment of spinal muscular atrophy. Discovery of this RT enzyme opens up the possibility to quantify the level and half-life of this drug inside patients, which could aid treatment.
The work was funded by UKRI MRC, BBSRC, and the AstraZeneca/LMB Blue Sky Initiative.
Further references
Discovery and evolution of RNA and XNA reverse transcriptase function and fidelity. Houlihan, G., Arangundy-Franklin, S., Porebski, BT., Subramanian, N., Taylor, AI., Holliger, P. Nature Chemistry (Epub ahead of print)
Philipp’s group page